心肌僵硬度的高级统计推断:模拟心脏力学的时间序列高斯过程方法,用于实时临床决策支持。
IF 7
2区 医学
Q1 BIOLOGY
引用次数: 0
摘要
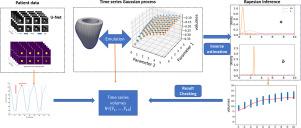
心脏力学建模有望彻底改变个性化医疗保健;然而,推断患者特定的生物物理参数对了解心肌功能和性能至关重要,这在方法论上提出了巨大挑战。我们的工作主要是通过测量不同时间点的左心室(LV)容积来确定心肌的被动刚度,这对诊断心脏生理状况至关重要。尽管在心脏力学建模方面取得了重大进展,但心肌僵硬度的推理和不确定性量化任务仍然具有挑战性,高昂的计算成本阻碍了实时决策支持。为了克服这一难题,我们采用高斯过程构建了一个统计代用模型,用于模拟舒张期充盈时的左心室腔容积。由于在舒张期不同时间点获得的左心室容积构成了一个时间序列,我们应用克朗内克乘积技巧将整个系统的复杂协方差矩阵分解为两个独立的协方差矩阵,一个是时间矩阵,另一个是生物物理参数矩阵。为了实现个性化医疗,我们利用主成分分析(PCA)进一步将患者特定的左心室几何形状整合到高斯过程模拟器中。利用深度学习神经网络从磁共振图像中提取时间序列左心室容积,并应用贝叶斯推理确定关键心脏力学参数的后验概率分布。对真实患者数据的测试表明,实时估算心肌特性可为临床决策提供帮助。这些进展是向临床影响迈出的关键一步,为复杂心脏力学模型的后验不确定性量化提供了宝贵的见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Advanced statistical inference of myocardial stiffness: A time series Gaussian process approach of emulating cardiac mechanics for real-time clinical decision support
Cardiac mechanics modelling promises to revolutionize personalized health care; however, inferring patient-specific biophysical parameters, which are critical for understanding myocardial functions and performance, poses substantial methodological challenges. Our work is primarily motivated to determine the passive stiffness of the myocardium from the measurement of the left ventricle (LV) volume at various time points, which is crucial for diagnosing cardiac physiological conditions. Although there have been significant advancements in cardiac mechanics modelling, the tasks of inference and uncertainty quantification of myocardial stiffness remain challenging, with high computational costs preventing real-time decision support. We adapt Gaussian processes to construct a statistical surrogate model for emulating LV cavity volume during diastolic filling to overcome this challenge. As the LV volumes, obtained at different time points in diastole, constitute a time series, we apply the Kronecker product trick to decompose the complex covariance matrix of the whole system into two separate covariance matrices, one for time and the other for biophysical parameters. To proceed towards personalized health care, we further integrate patient-specific LV geometries into the Gaussian process emulator using principal component analysis (PCA). Utilizing a deep learning neural network for extracting time-series left ventricle volumes from magnetic resonance images, Bayesian inference is applied to determine the posterior probability distribution of critical cardiac mechanics parameters. Tests on real-patient data illustrate the potential for real-time estimation of myocardial properties for clinical decision-making. These advancements constitute a crucial step towards clinical impact, offering valuable insights into posterior uncertainty quantification for complex cardiac mechanics models.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


