通过免疫信息学方法设计多表位腐肉病候选疫苗。
IF 7
2区 医学
Q1 BIOLOGY
引用次数: 0
摘要
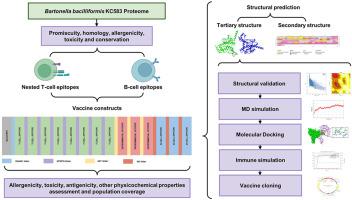
由巴氏杆菌引起的卡里翁病是秘鲁、厄瓜多尔和哥伦比亚的一个严重公共卫生问题。目前还没有针对巴氏杆菌的疫苗。虽然抗生素是标准的治疗方法,但耐药菌株已有报道,而且传播细菌的病媒也有可能扩散。本研究旨在利用免疫信息学工具设计一种针对卡里昂氏病病原体的多表位候选疫苗。研究人员利用秘鲁、厄瓜多尔、哥伦比亚和全球最常见的等位基因,从杆菌 KC583 的整个蛋白质组中预测了 B 细胞表位以及 CD4+ 和 CD8+T 细胞表位。从外膜蛋白和毒力相关蛋白中筛选出了 B 细胞表位和 T 细胞嵌套表位。表位是根据杂合性、非致敏性、保护性、非同源性和无毒性筛选出来的。使用连接体组装了两个疫苗构建体。对构建体的三级结构进行了预测,并通过分子动力学模拟对其稳定性进行了评估。选择了最稳定的构建物与 TLR4 受体进行分子对接。本研究提出了一种疫苗构建体,并对其进行了硅学评估,认为它是一种潜在的巴氏杆菌候选疫苗。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Design of a multi-epitope vaccine candidate against carrion disease by immunoinformatics approach
Carrion's disease, caused by the bacterium Bartonella bacilliformis, is a serious public health problem in Peru, Ecuador and Colombia. Currently there is no available vaccine against B. bacilliformis. While antibiotics are the standard treatment, resistant strains have been reported, and there is a potential spread of the vector that transmits the bacteria. This study aimed to design a multi-epitope vaccine candidate against the causative agent of Carrion's disease using immunoinformatics tools. Predictions of B-cell epitopes, as well as CD4+ and CD8+T cell epitopes, were performed from the entire proteome of B. bacilliformis KC583 using the most frequent alleles from Peru, Ecuador, Colombia, and worldwide. B-cell epitopes and T-cell nested epitopes from outer membrane and virulence-associated proteins were selected. Epitopes were filtered out based on promiscuity, non-allergenicity, conservation, non-homology and non-toxicity. Two vaccine constructs were assembled using linkers. The tertiary structure of the constructs was predicted, and their stability was evaluated through molecular dynamics simulations. The most stable construct was selected for molecular docking with the TLR4 receptor. This study proposes a vaccine construct evaluated in silico as a potential vaccine candidate against Bartonella bacilliformis.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


