以混合深度学习为指导的生成式人工智能重塑自组装肽的发现过程
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
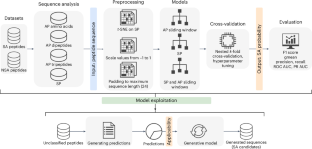
超分子肽基材料在纳米技术和医学等领域具有巨大的变革潜力。然而,破译其实际应用所必需的复杂序列到组装途径仍然是一项具有挑战性的工作。它们的发现主要依靠经验方法,需要大量的财政资源,这阻碍了它们的颠覆性潜力。因此,尽管自组装肽的特征繁多且优势明显,但只有少数肽材料进入了市场。根据实验验证数据训练的机器学习是一种很有前途的工具,可用于快速识别具有高度自组装倾向的序列,从而将资源支出集中在最有前途的候选产品上。在这里,我们介绍了一个框架,该框架在基于元启发式的生成模型中实施了精确的分类器,以引导在具有挑战性大小的肽序列空间中进行搜索。为此,我们训练了五个递归神经网络,其中使用聚集倾向和特定理化性质序列信息的混合模型取得了卓越的性能,准确率达到 81.9%,F1 分数为 0.865。分子动力学模拟和实验验证证实,该生成模型在发现自组装肽方面的准确率为 80-95%,优于目前最先进的模型。在探索自组装肽的过程中,所提出的模块化框架有效地补充了人类的直觉,为开发加速材料发现的智能实验室迈出了重要一步。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Reshaping the discovery of self-assembling peptides with generative AI guided by hybrid deep learning
Supramolecular peptide-based materials have great potential for revolutionizing fields like nanotechnology and medicine. However, deciphering the intricate sequence-to-assembly pathway, essential for their real-life applications, remains a challenging endeavour. Their discovery relies primarily on empirical approaches that require substantial financial resources, impeding their disruptive potential. Consequently, despite the multitude of characterized self-assembling peptides and their demonstrated advantages, only a few peptide materials have found their way to the market. Machine learning trained on experimentally verified data presents a promising tool for quickly identifying sequences with a high propensity to self-assemble, thereby focusing resource expenditures on the most promising candidates. Here we introduce a framework that implements an accurate classifier in a metaheuristic-based generative model to navigate the search through the peptide sequence space of challenging size. For this purpose, we trained five recurrent neural networks among which the hybrid model that uses sequential information on aggregation propensity and specific physicochemical properties achieved a superior performance with 81.9% accuracy and 0.865 F1 score. Molecular dynamics simulations and experimental validation have confirmed the generative model to be 80–95% accurate in the discovery of self-assembling peptides, outperforming the current state-of-the-art models. The proposed modular framework efficiently complements human intuition in the exploration of self-assembling peptides and presents an important step in the development of intelligent laboratories for accelerated material discovery. A generative model guided by a machine-learning-based classifier capable of assessing unexplored regions of the peptide space in the search for new self-assembling sequences.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


