了解乳腺癌异种移植模型中肿瘤生长的可变性,确定 PARP 抑制抗性生物标记物。
IF 6.8
1区 医学
Q1 ONCOLOGY
引用次数: 0
摘要
了解 PARP 抑制剂(PARPi)的耐药机制是临床的当务之急,尤其是在乳腺癌领域。我们开发了一种新的数学框架,通过将模型与乳腺癌患者异种移植(PDX)数据中的肿瘤生长指标相拟合来确定奥拉帕利的内在耐药性。治疗前的转录组图谱与计算出的耐药性一起用于确定耐药性的基线生物标志物,包括潜在的联合靶点。该模型既提供了反应分类,也提供了耐药性的连续描述,使生物标志物关联更加稳健,并捕捉到了观察到的变异性。研究发现了 36 个耐药性基因标记,包括多个同源重组修复 (HRR) 通路基因。WEE1 的高表达也与耐药性有关,这凸显了结合使用 PARP 和 WEE1 抑制剂的机会。该框架有助于以全自动的方式捕捉内在耐药性,并考虑到了PDX研究中捕捉到的药理反应变异性,从而提供了一种精准医疗方法。本文章由计算机程序翻译,如有差异,请以英文原文为准。
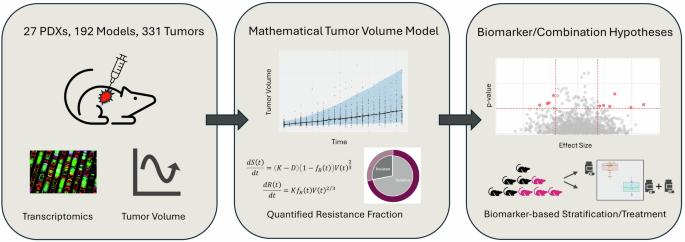
Understanding tumour growth variability in breast cancer xenograft models identifies PARP inhibition resistance biomarkers
Understanding the mechanisms of resistance to PARP inhibitors (PARPi) is a clinical priority, especially in breast cancer. We developed a novel mathematical framework accounting for intrinsic resistance to olaparib, identified by fitting the model to tumour growth metrics from breast cancer patient-derived xenograft (PDX) data. Pre-treatment transcriptomic profiles were used with the calculated resistance to identify baseline biomarkers of resistance, including potential combination targets. The model provided both a classification of responses, as well as a continuous description of resistance, allowing for more robust biomarker associations and capturing the observed variability. Thirty-six resistance gene markers were identified, including multiple homologous recombination repair (HRR) pathway genes. High WEE1 expression was also linked to resistance, highlighting an opportunity for combining PARP and WEE1 inhibitors. This framework facilitates a fully automated way of capturing intrinsic resistance, and accounts for the pharmacological response variability captured within PDX studies and hence provides a precision medicine approach.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

NPJ Precision Oncology
ONCOLOGY-
CiteScore
9.90
自引率
1.30%
发文量
87
审稿时长
18 weeks
期刊介绍:
Online-only and open access, npj Precision Oncology is an international, peer-reviewed journal dedicated to showcasing cutting-edge scientific research in all facets of precision oncology, spanning from fundamental science to translational applications and clinical medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


