系统发生组学解析了带状蝾螈(Ommatotriton 属)令人费解的系统发生。
IF 3.6
1区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
解决快速连续发生的物种变异事件的顺序问题本身就很困难,通常需要采用系统发生学方法。带状蝾螈(Ommatotriton 属)三个物种的系统发育之前尚未解决,这就是一个很好的例子。我们通过序列捕获目标富集法获得了约 7k 个核 DNA 标记,并使用 RAxML 对连接数据进行最大似然推断,使用 ASTRAL 进行多物种聚合分析,使用 SNAPPER 的扩散模型进行贝叶斯物种树推断,并使用 TreeMix 和 PhyloNet 测试种间基因流。所有分析都恢复了三个不同的物种,没有证据表明存在种间基因流。所有分析都检索到了拓扑结构(O. nesterovi、(O. ophryticus、O. vittatus)),支持率很高。SNAPPER 确实显示出陷入局部最优的趋势,从而产生了不同的拓扑结构,但支持率仍然很高。此外,我们注意到,当我们包含一个外群时,陷入局部最优的 SNAPPER 运行次数较少。因此,我们建议使用这种方法探索多个独立运行并使用外群。带状蝾螈辐射说明了如何利用全基因组数据来解决以前悬而未决的系统发育问题。本文章由计算机程序翻译,如有差异,请以英文原文为准。
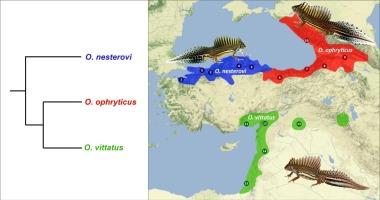
Phylogenomics resolves the puzzling phylogeny of banded newts (genus Ommatotriton)
Resolving the order of speciation events that occurred in rapid succession is inherently hard and typically requires a phylogenomic approach. A case in point concerns the previously unresolved phylogeny of the three species of banded newt (genus Ommatotriton). We obtain c. 7k nuclear DNA markers using target enrichment by sequence capture and analyze the dataset using maximum likelihood inference of concatenated data with RAxML, summary multi-species coalescent analysis with ASTRAL and Bayesian species tree inference using a diffusion model with SNAPPER, and use TreeMix and PhyloNet to test for interspecific gene flow. All analyses recover three distinct species with no evidence of interspecific gene flow. All analyses retrieved the topology (O. nesterovi, (O. ophryticus, O. vittatus)), with high support. SNAPPER did show the tendency to get stuck in a local optimum, resulting in a different but still highly supported topology. Furthermore, we notice that fewer SNAPPER runs get stuck in a local optimum when we include an outgroup. Therefore, we recommend the exploration of multiple independent runs and the use of an outgroup with this approach. The banded newt radiation illustrates the use of genome-wide data to tackle formerly unresolved phylogenies.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
7.50
自引率
7.30%
发文量
249
审稿时长
7.5 months
期刊介绍:
Molecular Phylogenetics and Evolution is dedicated to bringing Darwin''s dream within grasp - to "have fairly true genealogical trees of each great kingdom of Nature." The journal provides a forum for molecular studies that advance our understanding of phylogeny and evolution, further the development of phylogenetically more accurate taxonomic classifications, and ultimately bring a unified classification for all the ramifying lines of life. Phylogeographic studies will be considered for publication if they offer EXCEPTIONAL theoretical or empirical advances.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


