从两栖动物转录组数据中筛选新型抗菌素并计算其特征。
IF 2.6
4区 生物学
Q2 BIOLOGY
引用次数: 0
摘要
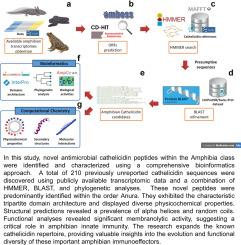
作为生活在潮湿和黑暗环境中的冷血生物,两栖动物进化出了强大的防御机制来保护自己免受捕食者和感染的侵害。在两栖动物产生的大量生物活性化合物中,抗菌肽(AMPs)是先天免疫所必需的。其中一类重要的 AMPs 是柔毛鞘氨醇,因其对病原体的广谱活性和免疫调节作用而闻名。然而,尽管它们具有广阔的生物医学潜力,而且全局数据的可用性也在不断提高,但对两栖动物中猫肝素的研究却寥寥无几,大多数研究都是通过传统的实验技术进行的。在这里,我们从两栖动物的转录组中发现了210个新的柔毛素序列,这些序列是通过综合计算管道确定的,计算管道采用了HMMER和BLAST工具来筛选柔毛素结构域。这些序列揭示了典型的三方结构域结构,并通过 SignalP 和 InterProScan 分析得到了证实。利用 IQ-TREE 进行的系统发育推断根据进化关系将这些序列分为六类。与其他脊椎动物的柔毛球蛋白相比,两栖动物的成熟肽平均长度较长(约 50 个氨基酸),芳香族和疏水残基较少,热稳定性较低。此外,对这些两栖动物柔毛鞘氨醇的理化和生物学特性进行了表征,发现它们具有显著的抗菌潜力,但溶血能力较低,尤其是在无尾目动物中,这表明通过 AMPlify、ampir、AmpGram 和 HemoPI 预测的抗菌和溶血活性之间存在平衡。二级结构估计(包括使用 AlphaFold2 进行的三维建模)表明,两栖动物的柔毛素主要以 α-螺旋和线圈为特征。一些具有代表性的模型还显示出具有两性拓扑结构的高α-螺旋组成,这有利于通过PPM方法评估与模拟细菌膜的相互作用。因此,这些发现突显了柔毛球蛋白在两栖动物免疫中的功能性作用及其良好的生物医学应用前景,强调了应用计算方法扩大范围和揭示脊椎动物柔毛球蛋白多样性景观的重要性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Screening and computational characterization of novel antimicrobial cathelicidins from amphibian transcriptomic data
As cold-blooded organisms living in damp and dark environments, amphibians have evolved robust defense mechanisms to protect themselves from predators and infections. Among the wide repertoire of bioactive compounds they produce are antimicrobial peptides (AMPs), which are required as part of innate immunity. One important class of AMPs is cathelicidins, known for their broad-spectrum activity against pathogens and their immunoregulatory roles. However, despite their promising biomedical potential and the increasing availability of omics data, few cathelicidins have been studied in amphibians, mostly through conventional experimental techniques. Here, we present 210 novel cathelicidin sequences from amphibian transcriptomes, identified through a comprehensive computational pipeline, which employed HMMER and BLAST tools to screen cathelicidin domains. These sequences reveal a typical tripartite domain architecture that was confirmed by SignalP and InterProScan analysis. Phylogenetic inference with IQ-TREE classified the sequences into six categories based on evolutionary relationships. Compared to cathelicidins from other vertebrates, amphibian mature peptides exhibit longer average lengths (around 50 amino acids), fewer aromatic and hydrophobic residues, and reduced thermal stability. Furthermore, these amphibian cathelicidins were characterized for their physicochemical and biological properties, revealing significant antimicrobial potential with lower hemolytic capability, especially in anurans, which suggests a balance between their antimicrobial and hemolytic activities predicted through AMPlify, ampir, AmpGram, and HemoPI. Secondary structure estimations, including three-dimensional modeling using AlphaFold2, indicate that amphibian cathelicidins predominantly feature -helices and coils. Some representative models also display a high -helix composition with amphipathic topology, facilitating interactions with simulated bacterial membranes as assessed by the PPM approach. Thus, these findings highlight the functional role of cathelicidins in amphibian immunity and their promising biomedical applicability, emphasizing the importance of applying computational methods to expand the scope and reveal the diverse landscape of cathelicidins across vertebrates.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Computational Biology and Chemistry
生物-计算机:跨学科应用
CiteScore
6.10
自引率
3.20%
发文量
142
审稿时长
24 days
期刊介绍:
Computational Biology and Chemistry publishes original research papers and review articles in all areas of computational life sciences. High quality research contributions with a major computational component in the areas of nucleic acid and protein sequence research, molecular evolution, molecular genetics (functional genomics and proteomics), theory and practice of either biology-specific or chemical-biology-specific modeling, and structural biology of nucleic acids and proteins are particularly welcome. Exceptionally high quality research work in bioinformatics, systems biology, ecology, computational pharmacology, metabolism, biomedical engineering, epidemiology, and statistical genetics will also be considered.
Given their inherent uncertainty, protein modeling and molecular docking studies should be thoroughly validated. In the absence of experimental results for validation, the use of molecular dynamics simulations along with detailed free energy calculations, for example, should be used as complementary techniques to support the major conclusions. Submissions of premature modeling exercises without additional biological insights will not be considered.
Review articles will generally be commissioned by the editors and should not be submitted to the journal without explicit invitation. However prospective authors are welcome to send a brief (one to three pages) synopsis, which will be evaluated by the editors.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


