活细胞中的 TGF-β 受体特异性 NanoBRET 靶标参与,用于高通量激酶抑制剂筛选。
IF 2.7
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
靶向转化生长因子-β(TGF-β)受体是一种很有前景的药理学方法,可使与 TGF-β 相关的遗传性和非遗传性疾病(包括纤维化、癌症、心血管疾病和肌肉骨骼疾病)中的异常信号转导正常化。为鉴定新型 TGF-β 受体激酶抑制剂,通常采用体外激酶测定、Western 印迹或转录报告测定等方法进行筛选。虽然这些方法有一定的优势,但缺乏对受体特异性、高通量能力和细胞环境相似性等关键特征的整合仍是其主要缺点。这一缺陷最终会阻碍研究成果转化到药物开发的后期(临床)阶段。在本研究中,我们介绍了一种经过调整和优化的基于活细胞 NanoBRET Target Engagement (TE) 的方法,用于鉴定 TGF-β 受体特异性激酶抑制剂。这个综合工具包包含各种 TGF-β I 型和 II 型受体、相应的 nanoBRET 示踪剂以及与疾病相关的细胞系,包括新型非商业性材料。当使用稳定表达的细胞系和低浓度示踪剂时,纳米BRET能力和激酶抑制窗口可显著增强功能测量。此外,该系统还可定制用于研究与 TGF-β 相关的遗传疾病,并可能用于筛选疾病特异性疗法。因此,在未来针对 TGF-β/BMP 受体的高通量化合物筛选中,我们非常鼓励使用这种优化的、活细胞的、不依赖抗体的 nanoBRET 靶点接合测定。本文章由计算机程序翻译,如有差异,请以英文原文为准。
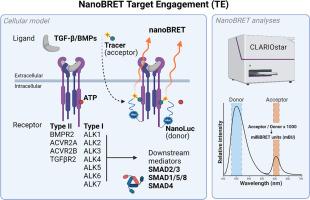
TGF-β receptor-specific NanoBRET Target Engagement in living cells for high-throughput kinase inhibitor screens
Targeting transforming growth factor-β (TGF-β) receptors is a promising pharmacological approach to normalize aberrant signaling in genetic and non-genetic TGF-β associated diseases including fibrosis, cancer, cardiovascular and musculoskeletal disorders. To identify novel TGF-β receptor kinase inhibitors, methods like in vitro kinase assays, western blot or transcriptional reporter assays are often used for screening purposes. While these methods may have certain advantages, the lack of integration of key features such as receptor specificity, high-throughput capability, and cellular context resemblance remains a major disadvantage. This deficiency could ultimately hinder the translation of study outcomes into later (clinical) stages of drug development. In this study, we introduce an adjusted and optimized live cell NanoBRET Target Engagement (TE)-based method to identify TGF-β receptor specific kinase inhibitors. This comprehensive toolkit contains various TGF-β type I and type II receptors, with corresponding nanoBRET tracers, and disease-related cell lines, including novel non-commercially available materials. The nanoBRET capacity and kinase inhibitory window can be significantly enhanced for functional measurements when stable expression cell lines and substantially low tracer concentrations are used. In addition, this system can be tailored to study TGF-β associated genetic disorders and possibly be used to screen for disease-specific therapeutics. Therefore, the use of this optimized, live cell, antibody-independent nanoBRET Target Engagement assay is highly encouraged for future high-throughput compound screens targeting TGF-β/BMP receptors.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

SLAS Discovery
Chemistry-Analytical Chemistry
CiteScore
7.00
自引率
3.20%
发文量
58
审稿时长
39 days
期刊介绍:
Advancing Life Sciences R&D: SLAS Discovery reports how scientists develop and utilize novel technologies and/or approaches to provide and characterize chemical and biological tools to understand and treat human disease.
SLAS Discovery is a peer-reviewed journal that publishes scientific reports that enable and improve target validation, evaluate current drug discovery technologies, provide novel research tools, and incorporate research approaches that enhance depth of knowledge and drug discovery success.
SLAS Discovery emphasizes scientific and technical advances in target identification/validation (including chemical probes, RNA silencing, gene editing technologies); biomarker discovery; assay development; virtual, medium- or high-throughput screening (biochemical and biological, biophysical, phenotypic, toxicological, ADME); lead generation/optimization; chemical biology; and informatics (data analysis, image analysis, statistics, bio- and chemo-informatics). Review articles on target biology, new paradigms in drug discovery and advances in drug discovery technologies.
SLAS Discovery is of particular interest to those involved in analytical chemistry, applied microbiology, automation, biochemistry, bioengineering, biomedical optics, biotechnology, bioinformatics, cell biology, DNA science and technology, genetics, information technology, medicinal chemistry, molecular biology, natural products chemistry, organic chemistry, pharmacology, spectroscopy, and toxicology.
SLAS Discovery is a member of the Committee on Publication Ethics (COPE) and was published previously (1996-2016) as the Journal of Biomolecular Screening (JBS).
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


