利用深度学习对胰腺进行大规模多中心 CT 和 MRI 分割。
IF 10.7
1区 医学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
胰腺疾病的诊断和随访需要在横断面成像上对胰腺进行自动容积分割。虽然基于 CT 的胰腺分割方法已较为成熟,但基于 MRI 的分割方法还未得到充分研究,这主要是由于缺乏公开可用的数据集、基准研究工作以及特定领域的深度学习方法。在这项回顾性研究中,我们收集了 2004 年 3 月至 2022 年 11 月期间来自五个中心的 T1 加权(T1 W)和 T2 加权(T2 W)腹部 MRI 系列的大型数据集(来自 499 名参与者的 767 次扫描)。我们还从公开来源收集了 1,350 名患者的 CT 扫描结果,以作为基准。我们引入了一种新的胰腺分割方法,称为 PanSegNet,它结合了 nnUNet 和 Transformer 网络的优势,并采用了新的线性注意模块,实现了体积计算。我们使用 Dice 和 Hausdorff 距离 (HD95) 评估指标测试了 PanSegNet 在跨模态(共 2,117 次扫描)和跨中心设置下的准确性。我们使用 Cohen's kappa 统计法对评分者内部和评分者之间的一致性进行评估,并使用配对 t 检验法分别对体积和 Dice 进行比较。在分割准确性方面,CT 的 Dice 系数为 88.3%(±7.2%,病例水平),T1 W MRI 为 85.0%(±7.9%),T2 W MRI 为 86.3%(±6.4%)。胰腺体积预测的相关性很高,CT、T1 W 和 T2 W 的 R2 分别为 0.91、0.84 和 0.85。我们发现观察者之间的一致性得分中等(T1 W 和 T2 W MRI 的一致性得分分别为 0.624 和 0.638),观察者内部的一致性得分较高。所有 MRI 数据可在 https://osf.io/kysnj/ 网站上查阅。我们的源代码见 https://github.com/NUBagciLab/PaNSegNet。本文章由计算机程序翻译,如有差异,请以英文原文为准。
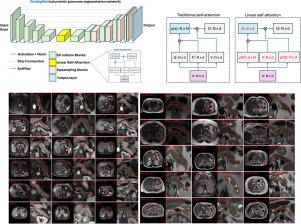
Large-scale multi-center CT and MRI segmentation of pancreas with deep learning
Automated volumetric segmentation of the pancreas on cross-sectional imaging is needed for diagnosis and follow-up of pancreatic diseases. While CT-based pancreatic segmentation is more established, MRI-based segmentation methods are understudied, largely due to a lack of publicly available datasets, benchmarking research efforts, and domain-specific deep learning methods. In this retrospective study, we collected a large dataset (767 scans from 499 participants) of T1-weighted (T1 W) and T2-weighted (T2 W) abdominal MRI series from five centers between March 2004 and November 2022. We also collected CT scans of 1,350 patients from publicly available sources for benchmarking purposes. We introduced a new pancreas segmentation method, called PanSegNet, combining the strengths of nnUNet and a Transformer network with a new linear attention module enabling volumetric computation. We tested PanSegNet’s accuracy in cross-modality (a total of 2,117 scans) and cross-center settings with Dice and Hausdorff distance (HD95) evaluation metrics. We used Cohen’s kappa statistics for intra and inter-rater agreement evaluation and paired t-tests for volume and Dice comparisons, respectively. For segmentation accuracy, we achieved Dice coefficients of 88.3% (±7.2%, at case level) with CT, 85.0% (±7.9%) with T1 W MRI, and 86.3% (±6.4%) with T2 W MRI. There was a high correlation for pancreas volume prediction with of 0.91, 0.84, and 0.85 for CT, T1 W, and T2 W, respectively. We found moderate inter-observer (0.624 and 0.638 for T1 W and T2 W MRI, respectively) and high intra-observer agreement scores. All MRI data is made available at https://osf.io/kysnj/. Our source code is available at https://github.com/NUBagciLab/PaNSegNet.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Medical image analysis
工程技术-工程:生物医学
CiteScore
22.10
自引率
6.40%
发文量
309
审稿时长
6.6 months
期刊介绍:
Medical Image Analysis serves as a platform for sharing new research findings in the realm of medical and biological image analysis, with a focus on applications of computer vision, virtual reality, and robotics to biomedical imaging challenges. The journal prioritizes the publication of high-quality, original papers contributing to the fundamental science of processing, analyzing, and utilizing medical and biological images. It welcomes approaches utilizing biomedical image datasets across all spatial scales, from molecular/cellular imaging to tissue/organ imaging.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


