通过计算发现针对 5'- 甲基硫代腺苷/S-腺苷高半胱氨酸核苷酸酶(MTAN)的 AI-2 法定量感应抑制剂,以抗击耐药性幽门螺旋杆菌。
IF 7
2区 医学
Q1 BIOLOGY
引用次数: 0
摘要
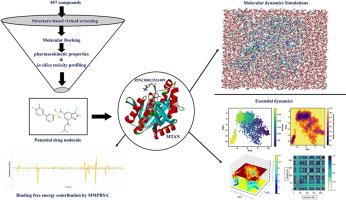
MTAN是幽门螺杆菌的一个可实现的治疗靶点,因为它可以在不影响肠道菌群的情况下最大限度地减少毒力产生、限制抗药性并损害法定量感应。本文对 457 种具有抗幽门螺杆菌活性的化合物进行了方法学分析,发现了一系列不同的化学类别和独特的化合物。分子对接研究发现了三种具有高结合亲和力的潜在化合物,即去氢木香内酯、角胺 B 和 ZINC00013531409,它们的结合亲和力分别为 -7.9、-9.2 和 -8.3 kcal/mol。与 Apo-MTAN 相比,ZINC00013531409-MTAN 相互作用的分子动力学模拟显示了 300 ns 的稳定性和相互作用,并阐明了参与蛋白质配体结合的关键残基。对氢键(Ile52、Met174 和 Arg194)和二级结构变化的分析进一步阐明了复合物内部的结合相互作用和构象变化。结合自由能计算揭示了支配 ZINC00013531409-MTAN 复合物形成的能量和相互作用。PCA 阐明了主要的运动模式,FEL 揭示了能量上有利的状态,然后 DCCM 揭示了残基之间的相关运动。总之,本研究对具有抗幽门螺杆菌活性的 ZINC00013531409 进行了详细的计算评估,突出了其毒性特征、构象稳定性和结合相互作用,为进一步开发抗菌药物奠定了基础。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Computational-driven discovery of AI-2 quorum sensing inhibitor targeting the 5′- methylthioadenosine/S-adenosylhomocysteine nucleosidase (MTAN) to combat drug-resistant Helicobacter pylori
MTAN is an attainable therapeutic target for H. pylori because it may minimize virulence production, limit resistance, and impair quorum sensing without affecting gut flora. Here, 457 compounds with anti-H. pylori activity were methodically analyzed, revealing a diverse array of chemical classes and unique compounds. Molecular docking studies identified three potential compounds with high binding affinities, Dehydrocostus lactone, keramamine B, and ZINC00013531409, each having binding affinity of −7.9, −9.2, and −8.3 kcal/mol, respectively. Molecular dynamics simulations of the ZINC00013531409-MTAN interactions in comparison with Apo-MTAN demonstrated stability and interactions of 300 ns, with key residues involved in protein-ligand binding illuminated. Analysis of hydrogen bonds (Ile52, Met174, and Arg194) and secondary structure variations further elucidated the binding interactions and conformational changes within the complex. Binding free energy calculations shed light on the energetics and interactions governing the complex formation of the ZINC00013531409-MTAN complex. PCA elucidated the dominant modes of motion, along with FEL revealed the energetically favorable states and then DCCM shed light on the correlated motions between residues. Overall, this study offers a detailed computational evaluation of ZINC00013531409 with anti-H. pylori activity, highlighting toxicity profile, conformational stability, and binding interactions, providing a foundation for further drug development efforts toward bacterial resistance.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


