SARS-CoV-2 主要蛋白酶(M-pro)突变分析:洞察突变冷门
IF 6.3
2区 医学
Q1 BIOLOGY
引用次数: 0
摘要
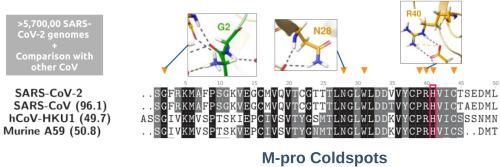
SARS-CoV-2 和 COVID-19 大流行标志着世界科学研究史上的一个里程碑。为了确保治疗在中长期内取得成功,确定 SARS-CoV-2 基因突变的特征至关重要,因为这些突变可能导致病毒耐药性。我们利用 GISAID 现有的超过 5,700,000 个 SARS-CoV-2 基因组的数据来报告 SARS-CoV-2 基因突变。鉴于其主要蛋白酶(M-pro)在病毒复制中的关键作用,我们对 SARS-CoV-2 M-pro 变异进行了详细分析,并特别关注抗突变残基或突变冷点,即在五个或更少的基因组中发生突变的残基。共发现了 32 个突变冷点,其中大部分介导了原体间的相互作用或从底物结合位点到二聚化表面的漏斗状相互作用网络,反之亦然。此外,突变冷点在其他 CoVs 的所有主要蛋白酶中几乎都是保守的。我们的研究结果提供了有关M-pro结构关键残基的宝贵信息,这些信息可能有助于合理的靶向药物设计,并为基于突变分析的M-pro二聚化抑制奠定了坚实的基础,有可能适用于未来的冠状病毒爆发。本文章由计算机程序翻译,如有差异,请以英文原文为准。
SARS-CoV-2 main protease (M-pro) mutational profiling: An insight into mutation coldspots
SARS-CoV-2 and the COVID-19 pandemic have marked a milestone in the history of scientific research worldwide. To ensure that treatments are successful in the mid-long term, it is crucial to characterize SARS-CoV-2 mutations, as they might lead to viral resistance. Data from >5,700,000 SARS-CoV-2 genomes available at GISAID was used to report SARS-CoV-2 mutations. Given the pivotal role of its main protease (M-pro) in virus replication, a detailed analysis of SARS-CoV-2 M-pro mutations was conducted, with particular attention to mutation-resistant residues or mutation coldspots, defined as those residues that have mutated in five or fewer genomes. 32 mutation coldspots were identified, most of which mediate interprotomer interactions or funneling interaction networks from the substrate-binding site towards the dimerization surface and vice versa. Besides, mutation coldspots were virtually conserved in all main proteases from other CoVs. Our results provide valuable information about key residues to M-pro structure that could be useful in rational target-directed drug design and establish a solid groundwork based on mutation analyses for the inhibition of M-pro dimerization, with a potential applicability to future coronavirus outbreaks.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


