对分枝杆菌 NudC 的结构研究揭示了一类独立于锌的 NADH 焦磷酸酶。
IF 4.7
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
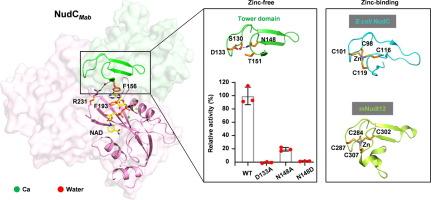
非结核分枝杆菌(NTM)对抗生素具有极强的耐药性,对公共卫生的威胁日益严重。有人提出,NADH焦磷酸酶(NudC)通过水解NAD修饰的活性形式(NAD-INH和NAD-ETH),参与分枝杆菌对一线抗结核药物异烟肼(INH)或其类似物乙硫酰胺(ETH)的耐药性。在本研究中,我们对来自脓肿霉菌的 NudC(NudCMab)进行了酶学和结构研究,脓肿霉菌对异烟肼高度耐药,正在成为最令人担忧的非典型肺炎霉菌。我们测定了 NudCMab 的蛋白酶、底物 NAD 结合型和产物 AMP 结合型晶体结构。我们观察到 NudCMab 的 Nudix 基序在三个二价阳离子的介导下捕获 NAD 的焦磷酸基团的模式,这为了解 NudC 水解 NAD(H) 或 NAD 封闭底物的机制提供了细节。有趣的是,我们的结构发现了一种来自分枝杆菌的新型亚类 NudC,其特点是在保守的 QPWPFPxS 基序上有一个独特的精氨酸残基,还有一个独特的塔状结构域,取代了大肠杆菌 NudC 中定义明确的锌结合基序和哺乳动物 Nudt12 的催化结构域。因此,我们对 NudCMab 的结构研究不仅展示了分枝杆菌中一类锌独立的 NADH 焦磷酸酶,还有助于设计 NudC 抑制剂,与 INH 或 ETH 联用治疗分枝杆菌感染。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Structural Studies on Mycobacterial NudC Reveal a Class of Zinc Independent NADH Pyrophosphatase
Non-tuberculous mycobacteria (NTM) have emerged as an increasing threat to public health, due to the extreme antibiotic resistance. NADH pyrophosphatase (NudC) was proposed involving in mycobacterial resistance to the first line anti-tubercular drug isoniazid (INH) or its analog ethionamide (ETH), by hydrolyzing their NAD modified active forms (NAD-INH and NAD-ETH). In this study, we performed enzymatic and structural studies on NudC from M. abscessus (NudCMab), which is highly resistant to isoniazid and emerging as the most worrisome NTM. We determined the crystal structures of NudCMab in apo form, substrate NAD-bound form and product AMP-bound form. We observed the mode for the Nudix motif of NudCMab capturing the pyrophosphate group of NAD mediated by three divalent cation ions, which provides details for understanding the mechanism on NudC hydrolyzing NAD(H) or NAD-capped substrate. Interestingly, our structures revealed a novel subclass NudC from mycobacteria characterized by a unique arginine residue on the conserved QPWPFPxS motif, as well as a unique tower domain that replaces a well-defined zinc-binding motif in E.coli NudC and catalytic domain of mammalian Nudt12. Thus, our structural studies on NudCMab not only present a class of zinc independent NADH pyrophosphatase in mycobacteria, but also may facilitate the design of NudC inhibitors for the treatment of mycobacteria infections in combination with INH or ETH.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


