验证 APOBEC3A 介导的 RNA 单碱基替换特征并提出新的 APOBEC1、APOBEC3B 和 APOBEC3G RNA 特征。
IF 4.7
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
突变特征分析为深入了解各种 DNA 单碱基置换(SBS)特征的潜在突变过程及其与不同癌症类型的关联提供了重要依据,因而备受关注。最近,通过分解在非小细胞肺癌中发现的 RNA 变异,定义并描述了 RNA 单碱基置换(RNA-SBS)特征。通过统计关联,他们将载脂蛋白 B mRNA 编辑酶催化多肽 3A(APOBEC3A)突变归因于 RNA-SBS2 特征。在此,我们通过将新型外源性和内源性 APOBEC3A RNA 编辑特征分解为 COSMICv3.4 RNA-SBS 参考特征,首次验证了 RNA-SBS 突变特征。此外,我们还发现了 APOBEC1、APOBEC3B 和 APOBEC3G 的新型 RNA-SBS 特征。本文章由计算机程序翻译,如有差异,请以英文原文为准。
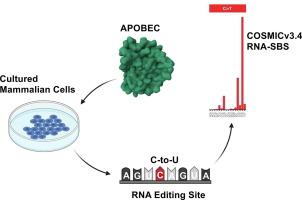
Validation of the APOBEC3A-Mediated RNA Single Base Substitution Signature and Proposal of Novel APOBEC1, APOBEC3B, and APOBEC3G RNA Signatures
Mutational signature analysis gained significant attention for providing critical insights into the underlying mutational processes for various DNA single base substitution (SBS) signatures and their associations with different cancer types. Recently, RNA single base substitution (RNA-SBS) signatures were defined and described by decomposing RNA variants found in non-small cell lung cancer. Through statistical association, they attributed Apolipoprotein B mRNA Editing Enzyme, Catalytic Polypeptide 3A (APOBEC3A) mutagenesis to the RNA-SBS2 signature. Here, we provide the first validation of an RNA-SBS mutational signature by decomposing novel exogenous and endogenous APOBEC3A RNA editing signatures into COSMICv3.4 RNA-SBS reference signatures. Additionally, we have identified novel RNA-SBS signatures for APOBEC1, APOBEC3B, and APOBEC3G.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


