从电子健康记录中提取儿科知识的多源表征学习
IF 12.4
1区 医学
Q1 HEALTH CARE SCIENCES & SERVICES
引用次数: 0
摘要
由于临床研究的高门槛,电子健康记录(EHR)系统对儿科尤为重要,但儿科 EHR 数据的内容密度往往较低。现有的电子病历代码嵌入是为普通患者量身定制的,无法满足儿科患者的独特需求。为了弥补这一缺陷,我们引入了一种迁移学习方法--MUltisource Graph Synthesis(MUGS),旨在儿科环境中进行准确的知识提取和关系检测。MUGS 整合了来自儿科和普通电子病历系统的图形数据以及分层医疗本体,创建了能自适应捕捉医院系统间同质性和异质性的嵌入。这些嵌入技术可实现完善的电子病历特征工程和细致入微的病人特征描述,在识别与特定特征相似的儿科病人(重点是肺动脉高压(PH))方面尤其有效。MUGS 嵌入抗负转移,在多种应用中优于其他基准方法,推动了循证儿科研究的发展。本文章由计算机程序翻译,如有差异,请以英文原文为准。
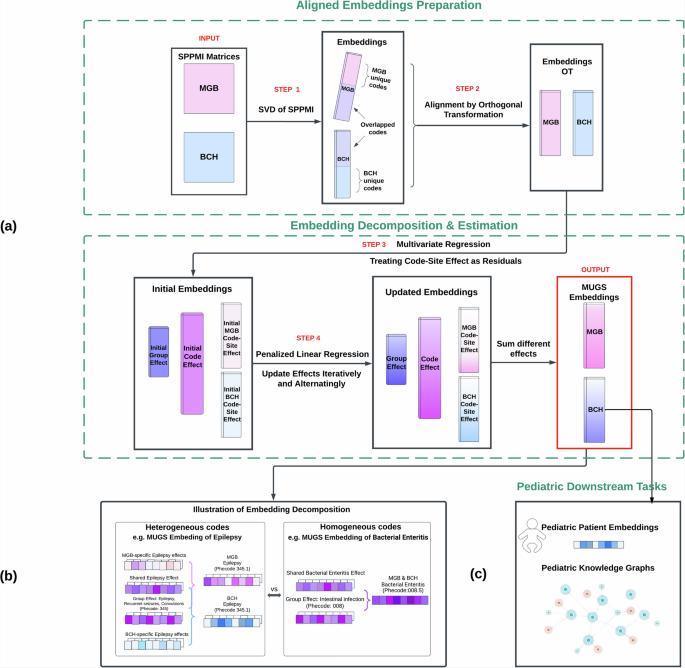
Multisource representation learning for pediatric knowledge extraction from electronic health records
Electronic Health Record (EHR) systems are particularly valuable in pediatrics due to high barriers in clinical studies, but pediatric EHR data often suffer from low content density. Existing EHR code embeddings tailored for the general patient population fail to address the unique needs of pediatric patients. To bridge this gap, we introduce a transfer learning approach, MUltisource Graph Synthesis (MUGS), aimed at accurate knowledge extraction and relation detection in pediatric contexts. MUGS integrates graphical data from both pediatric and general EHR systems, along with hierarchical medical ontologies, to create embeddings that adaptively capture both the homogeneity and heterogeneity between hospital systems. These embeddings enable refined EHR feature engineering and nuanced patient profiling, proving particularly effective in identifying pediatric patients similar to specific profiles, with a focus on pulmonary hypertension (PH). MUGS embeddings, resistant to negative transfer, outperform other benchmark methods in multiple applications, advancing evidence-based pediatric research.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

NPJ Digital Medicine
Multiple-
CiteScore
25.10
自引率
3.30%
发文量
170
审稿时长
15 weeks
期刊介绍:
npj Digital Medicine is an online open-access journal that focuses on publishing peer-reviewed research in the field of digital medicine. The journal covers various aspects of digital medicine, including the application and implementation of digital and mobile technologies in clinical settings, virtual healthcare, and the use of artificial intelligence and informatics.
The primary goal of the journal is to support innovation and the advancement of healthcare through the integration of new digital and mobile technologies. When determining if a manuscript is suitable for publication, the journal considers four important criteria: novelty, clinical relevance, scientific rigor, and digital innovation.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


