可重用性报告:探索变异图编码器在药物设计中预测分子毒性的实用性
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
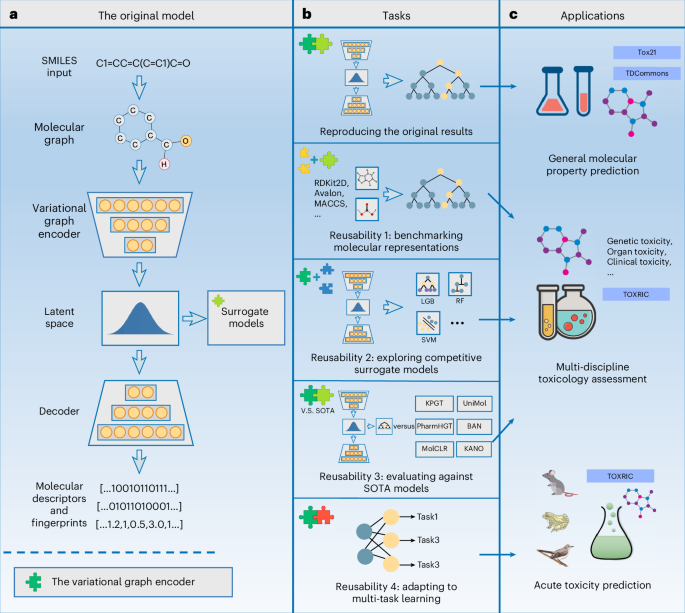
变异图编码器有效地结合了图卷积网络和变异自编码器,已被广泛用于生物医学图结构数据。Lam 及其同事开发了一个基于变异图编码器的框架 NYAN,以促进计算机辅助药物设计中的分子特性预测。在 NYAN 中,从变异图自动编码器中得到的低维潜在变量被用作通用的分子表示,在整个药物发现过程中产生了显著的性能和多功能性。在本研究中,我们评估了 NYAN 的可重用性,并研究了其在特定化学毒性预测中的适用性。基于 NYAN 潜在表征和其他常用分子特征表征的预测准确性在广泛的毒性数据集中进行了基准测试,同时还探讨了 NYAN 潜在表征对其他代用模型的适应性。与其他最先进的分子特性预测方法相比,配备了常用代用模型的 NYAN 在毒性预测方面具有竞争力或更好的性能。我们还利用 NYAN 潜在空间的低维度和特征多样性,设计了一种具有特征增强和共识推断功能的多任务学习策略,进一步提高了多端点急性毒性预测的能力。分析深入探讨了通用图变分法模型的适应性,展示了它在药物发现领域中执行定制任务的能力。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Reusability report: exploring the utility of variational graph encoders for predicting molecular toxicity in drug design
Variational graph encoders effectively combine graph convolutional networks with variational autoencoders, and have been widely employed for biomedical graph-structured data. Lam and colleagues developed a framework based on the variational graph encoder, NYAN, to facilitate the prediction of molecular properties in computer-assisted drug design. In NYAN, the low-dimensional latent variables derived from the variational graph autoencoder are leveraged as a universal molecular representation, yielding remarkable performance and versatility throughout the drug discovery process. In this study we assess the reusability of NYAN and investigate its applicability within the context of specific chemical toxicity prediction. The prediction accuracy—based on NYAN latent representations and other popular molecular feature representations—is benchmarked across a broad spectrum of toxicity datasets, and the adaptation of NYAN latent representation to other surrogate models is also explored. NYAN, equipped with common surrogate models, shows competitive or better performance in toxicity prediction compared with other state-of-the-art molecular property prediction methods. We also devise a multi-task learning strategy with feature enhancement and consensus inference by leveraging the low dimensionality and feature diversity of NYAN latent space, further boosting the multi-endpoint acute toxicity estimation. The analysis delves into the adaptability of the generic graph variational model, showcasing its aptitude for tailored tasks within the realm of drug discovery. Ruijiang Li et al. assess the reproducibility of a variational graph encoder-based framework and examines its reusability for chemical toxicity prediction. It explores how a generalist model can function as a specialist model with adaptation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


