用于识别 Cas9 上不同表位的纳米抗体工具箱。
IF 4.7
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
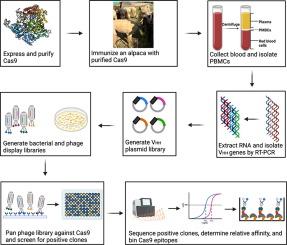
Cas9s 和 Cas9s 融合体已成为基因操作的强大工具。Cas9与其他DNA编辑酶的融合产生了能够进行单碱基编辑的变体,而催化死亡的Cas9则成为了特异性靶向基因组所需区域的工具。在这里,我们描述了针对化脓性链球菌 Cas9 上的三个独特表位生成纳米抗体的过程。这些纳米抗体是从用 Cas9 免疫的羊驼身上提取的纳米抗体库中鉴定出来的。最有效的结合体同样能识别Cas9和RNA结合的Cas9,而且不会抑制Cas9裂解靶DNA。根据基于酶联免疫吸附法的表位分选实验和质量光度测定法确定,这些纳米抗体结合的表位并不重叠。我们介绍了这些克隆的序列和支持性生化数据,以便更广泛的科学界能够获得这些试剂。本文章由计算机程序翻译,如有差异,请以英文原文为准。
A Nanobody Toolbox for Recognizing Distinct Epitopes on Cas9
Cas9s and fusions of Cas9s have emerged as powerful tools for genetic manipulations. Fusions of Cas9 with other DNA editing enzymes have led to variants capable of single base editing and catalytically dead Cas9s have emerged as tools to specifically target desired regions of a genome. Here we describe the generation of a panel of nanobodies directed against three unique epitopes on Streptococcus pyogenes Cas9. The nanobodies were identified from a nanobody library derived from an alpaca that had been immunized with Cas9. The most potent binders recognize Cas9 and RNA bound Cas9 equally well and do not inhibit Cas9 cleavage of target DNA. These nanobodies bind non-overlapping epitopes as determined by ELISA based epitope binning experiments and mass photometry. We present the sequences of these clones and supporting biochemical data so the broader scientific community can access these reagents.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


