通过蛋白-蛋白相互作用可解释图传播网络,基于血浆蛋白识别轻度认知障碍的神经影像驱动亚型
IF 7
2区 医学
Q1 BIOLOGY
引用次数: 0
摘要
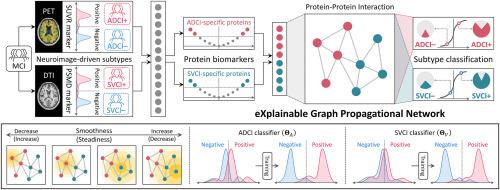
轻度认知障碍(MCI)是痴呆症的早期指标,需要根据其亚型进行专门治疗,以有效预防和控制痴呆症的发展。根据神经病理学特征,MCI 可分为阿尔茨海默病(AD)相关认知障碍(ADCI)和皮层下血管性认知障碍(SVCI),它们分别更有可能发展为 AD 和皮层下血管性痴呆(SVD)。在识别 MCI 亚型方面,血浆蛋白生物标记物因其在诊断过程中的微创性和成本效益,最近被视为很有前途的工具。此外,机器学习(ML)的应用提高了生物标记物发现和诊断的精确度。然而,以往基于 ML 的研究往往没有考虑到蛋白质之间的相互作用,而这种相互作用在 MCI 和痴呆症等复杂的神经退行性疾病中至关重要。虽然蛋白质-蛋白质相互作用(PPIs)已被应用于网络模型,但由于其局部意识,这些模型往往不能完全捕捉到 PPIs 的各种特性。这种局限性增加了忽略关键成分和放大嘈杂相互作用影响的可能性。在本研究中,我们引入了一种新的基于图的 ML 模型来对 MCI 亚型进行分类,该模型被称为 eXplainable Graph Propagational Network (XGPN)。该方法通过在 PPI 网络上传播血浆蛋白的独立效应来提取蛋白间的全局交互效应,从而通过估计每种蛋白的风险效应来预测 MCI 亚型。此外,由于 XGPN 架构的简单性和透明性,模型训练过程和亚型分类结果都是完全可解释的。实验结果表明,蛋白质之间的交互效应显著促进了 MCI 亚型组之间的明显差异,从而提高了分类性能,与现有方法相比平均提高了 10.0%,同时还识别了关键生物标志物及其对 ADCI 和 SVCI 的影响。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Plasma protein-based identification of neuroimage-driven subtypes in mild cognitive impairment via protein-protein interaction aware explainable graph propagational network
As an early indicator of dementia, mild cognitive impairment (MCI) requires specialized treatment according to its subtypes for the effective prevention and management of dementia progression. Based on the neuropathological characteristics, MCI can be classified into Alzheimer's disease (AD)-related cognitive impairment (ADCI) and subcortical vascular cognitive impairment (SVCI), being more likely to progress to AD and subcortical vascular dementia (SVD), respectively. For identifying MCI subtypes, plasma protein biomarkers are recently seen as promising tools due to their minimal invasiveness and cost-effectiveness in diagnostic procedures. Furthermore, the application of machine learning (ML) has led the preciseness in the biomarker discovery and the resulting diagnostics. Nevertheless, previous ML-based studies often fail to consider interactions between proteins, which are essential in complex neurodegenerative disorders such as MCI and dementia. Although protein-protein interactions (PPIs) have been employed in network models, these models frequently do not fully capture the diverse properties of PPIs due to their local awareness. This limitation increases the likelihood of overlooking critical components and amplifying the impact of noisy interactions. In this study, we introduce a new graph-based ML model for classifying MCI subtypes, called eXplainable Graph Propagational Network (XGPN). The proposed method extracts the globally interactive effects between proteins by propagating the independent effect of plasma proteins on the PPI network, and thereby, MCI subtypes are predicted by estimation of the risk effect of each protein. Moreover, the process of model training and the outcome of subtype classification are fully explainable due to the simplicity and transparency of XGPN's architecture. The experimental results indicated that the interactive effect between proteins significantly contributed to the distinct differences between MCI subtype groups, resulting in an enhanced classification performance with an average improvement of 10.0 % compared to existing methods, also identifying key biomarkers and their impact on ADCI and SVCI.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


