BioKGrapher:从生物医学文献中自动构建知识图谱的初步评估
IF 4.1
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
Computational and structural biotechnology journal
Pub Date : 2024-10-17
DOI:10.1016/j.csbj.2024.10.017
引用次数: 0
摘要
背景 生物医学文献的增长给知识的提取和结构化带来了挑战。知识图谱(KG)通过表示生物医学实体之间的关系提供了一种解决方案。然而,手工构建知识图谱耗费大量人力和时间,因此需要自动化方法。这项工作介绍了 BioKGrapher,这是一种利用大规模出版物数据自动构建知识图谱的工具,重点关注与特定医疗条件相关的生物医学概念。方法 BioKGrapher 管道从命名实体识别和链接(NER+NEL)开始,提取 PubMed 中的生物医学概念并将其规范化,将其映射到统一医学语言系统(UMLS)。利用库尔巴克-莱伯勒发散和局部频率平衡对提取的概念进行加权和重新排序。然后将这些概念整合到分层 KG 中,并使用 SNOMED CT 和 NCIt 等术语形成关系。结果 BioKGrapher 有效地将生成的概念与德国肿瘤学指南项目(GGPO)的临床实践指南相一致,F1 分数高达 0.6。在多标签分类中,使用 BioKGrapher 癌症特异性 KG 的适配器注入模型比非特异性 KG 的微观 F1 分数提高了 0.89 个百分点,比三种 BERT 变体的基础模型提高了 2.16 个百分点。药物-疾病提取案例研究确定了 Nivolumab 和 Rituximab 的适应症。它为管理生物医学知识提供了一个可扩展的解决方案,在文献推荐、决策支持和药物再利用方面具有潜在的应用价值。本文章由计算机程序翻译,如有差异,请以英文原文为准。
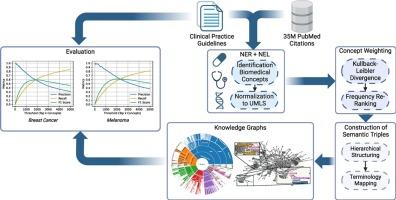
BioKGrapher: Initial evaluation of automated knowledge graph construction from biomedical literature
Background The growth of biomedical literature presents challenges in extracting and structuring knowledge. Knowledge Graphs (KGs) offer a solution by representing relationships between biomedical entities. However, manual construction of KGs is labor-intensive and time-consuming, highlighting the need for automated methods. This work introduces BioKGrapher, a tool for automatic KG construction using large-scale publication data, with a focus on biomedical concepts related to specific medical conditions. BioKGrapher allows researchers to construct KGs from PubMed IDs.
Methods The BioKGrapher pipeline begins with Named Entity Recognition and Linking (NER+NEL) to extract and normalize biomedical concepts from PubMed, mapping them to the Unified Medical Language System (UMLS). Extracted concepts are weighted and re-ranked using Kullback-Leibler divergence and local frequency balancing. These concepts are then integrated into hierarchical KGs, with relationships formed using terminologies like SNOMED CT and NCIt. Downstream applications include multi-label document classification using Adapter-infused Transformer models.
Results BioKGrapher effectively aligns generated concepts with clinical practice guidelines from the German Guideline Program in Oncology (GGPO), achieving -Scores of up to 0.6. In multi-label classification, Adapter-infused models using a BioKGrapher cancer-specific KG improved micro -Scores by up to 0.89 percentage points over a non-specific KG and 2.16 points over base models across three BERT variants. The drug-disease extraction case study identified indications for Nivolumab and Rituximab.
Conclusion BioKGrapher is a tool for automatic KG construction, aligning with the GGPO and enhancing downstream task performance. It offers a scalable solution for managing biomedical knowledge, with potential applications in literature recommendation, decision support, and drug repurposing.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Computational and structural biotechnology journal
Biochemistry, Genetics and Molecular Biology-Biophysics
CiteScore
9.30
自引率
3.30%
发文量
540
审稿时长
6 weeks
期刊介绍:
Computational and Structural Biotechnology Journal (CSBJ) is an online gold open access journal publishing research articles and reviews after full peer review. All articles are published, without barriers to access, immediately upon acceptance. The journal places a strong emphasis on functional and mechanistic understanding of how molecular components in a biological process work together through the application of computational methods. Structural data may provide such insights, but they are not a pre-requisite for publication in the journal. Specific areas of interest include, but are not limited to:
Structure and function of proteins, nucleic acids and other macromolecules
Structure and function of multi-component complexes
Protein folding, processing and degradation
Enzymology
Computational and structural studies of plant systems
Microbial Informatics
Genomics
Proteomics
Metabolomics
Algorithms and Hypothesis in Bioinformatics
Mathematical and Theoretical Biology
Computational Chemistry and Drug Discovery
Microscopy and Molecular Imaging
Nanotechnology
Systems and Synthetic Biology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


