PRISM:使用大型语言模型的病人记录语义临床试验匹配系统。
IF 12.4
1区 医学
Q1 HEALTH CARE SCIENCES & SERVICES
引用次数: 0
摘要
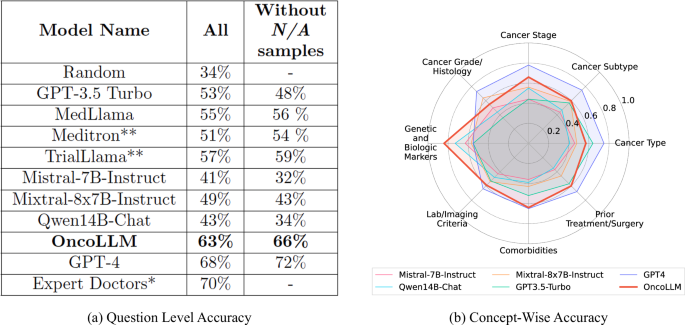
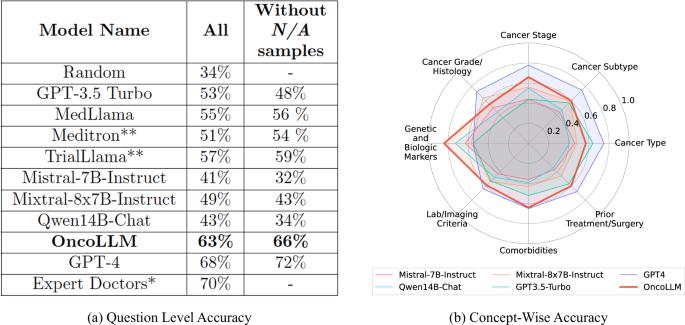
临床试验匹配是指确定患者可能符合条件的试验。通常情况下,这项工作需要耗费大量人力,而且需要根据临床试验严格的纳入和排除标准对患者的电子健康记录(EHR)进行详细核查。这一过程也导致许多患者错过了潜在的治疗方案。大语言模型(LLMs)的最新进展使患者与试验匹配的自动化成为可能,这一点已在多项并行研究中得到证实。然而,目前的方法仅限于受限数据集,通常是合成数据集,不能充分反映真实世界医疗数据中遇到的复杂性。在本研究中,我们对临床试验匹配系统进行了端到端的大规模实证评估,并使用真实世界的电子病历对其进行了验证。我们使用专有的 LLM 和我们定制的微调模型 OncoLLM 进行了全面的实验,结果表明 OncoLLM 的性能优于 GPT-3.5,并与合格医生的临床试验匹配性能相匹配。本文章由计算机程序翻译,如有差异,请以英文原文为准。
PRISM: Patient Records Interpretation for Semantic clinical trial Matching system using large language models
Clinical trial matching is the task of identifying trials for which patients may be eligible. Typically, this task is labor-intensive and requires detailed verification of patient electronic health records (EHRs) against the stringent inclusion and exclusion criteria of clinical trials. This process also results in many patients missing out on potential therapeutic options. Recent advancements in Large Language Models (LLMs) have made automating patient-trial matching possible, as shown in multiple concurrent research studies. However, the current approaches are confined to constrained, often synthetic, datasets that do not adequately mirror the complexities encountered in real-world medical data. In this study, we present an end-to-end large-scale empirical evaluation of a clinical trial matching system and validate it using real-world EHRs. We perform comprehensive experiments with proprietary LLMs and our custom fine-tuned model called OncoLLM and show that OncoLLM outperforms GPT-3.5 and matches the performance of qualified medical doctors for clinical trial matching.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

NPJ Digital Medicine
Multiple-
CiteScore
25.10
自引率
3.30%
发文量
170
审稿时长
15 weeks
期刊介绍:
npj Digital Medicine is an online open-access journal that focuses on publishing peer-reviewed research in the field of digital medicine. The journal covers various aspects of digital medicine, including the application and implementation of digital and mobile technologies in clinical settings, virtual healthcare, and the use of artificial intelligence and informatics.
The primary goal of the journal is to support innovation and the advancement of healthcare through the integration of new digital and mobile technologies. When determining if a manuscript is suitable for publication, the journal considers four important criteria: novelty, clinical relevance, scientific rigor, and digital innovation.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


