Jonathan E. Bravo, Ilya Slizovskiy, Nathalie Bonin, Marco Oliva, Noelle Noyes, Christina Boucher
{"title":"高灵敏度检测复杂微生物群落中 ARG-MGE 共定位的 TELCoMB 方案。","authors":"Jonathan E. Bravo, Ilya Slizovskiy, Nathalie Bonin, Marco Oliva, Noelle Noyes, Christina Boucher","doi":"10.1002/cpz1.70031","DOIUrl":null,"url":null,"abstract":"<p>Understanding the genetic basis of antimicrobial resistance is crucial for developing effective mitigation strategies. One necessary step is to identify the antimicrobial resistance genes (ARGs) within a microbial population, referred to as the resistome, as well as the mobile genetic elements (MGEs) harboring ARGs. Although shotgun metagenomics has been successful in detecting ARGs and MGEs within a microbiome, it is limited by low sensitivity. Enrichment using cRNA biotinylated probes has been applied to address this limitation, enhancing the detection of rare ARGs and MGEs, especially when combined with long-read sequencing. Here, we present the TELCoMB protocol, a Snakemake workflow that elucidates resistome and mobilome composition and diversity and uncovers ARG-MGE colocalizations. The protocol supports both short- and long-read sequencing and does not require enrichment, making it versatile for various genomic data types. TELCoMB generates publication-ready figures and CSV files for comprehensive analysis, improving our understanding of antimicrobial resistance mechanisms and spread. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Installing TELCOMB Locally</p><p><b>Alternate Protocol</b>: Installing TELCOMB on a SLURM Cluster</p><p><b>Basic Protocol 2</b>: Data Preprocessing</p><p><b>Basic Protocol 3</b>: Calculation of Resistome Distribution and Composition</p><p><b>Basic Protocol 4</b>: Identification of ARG-MGE Colocalizations</p>","PeriodicalId":93970,"journal":{"name":"Current protocols","volume":"4 10","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2024-10-24","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.70031","citationCount":"0","resultStr":"{\"title\":\"The TELCoMB Protocol for High-Sensitivity Detection of ARG-MGE Colocalizations in Complex Microbial Communities\",\"authors\":\"Jonathan E. Bravo, Ilya Slizovskiy, Nathalie Bonin, Marco Oliva, Noelle Noyes, Christina Boucher\",\"doi\":\"10.1002/cpz1.70031\",\"DOIUrl\":null,\"url\":null,\"abstract\":\"<p>Understanding the genetic basis of antimicrobial resistance is crucial for developing effective mitigation strategies. One necessary step is to identify the antimicrobial resistance genes (ARGs) within a microbial population, referred to as the resistome, as well as the mobile genetic elements (MGEs) harboring ARGs. Although shotgun metagenomics has been successful in detecting ARGs and MGEs within a microbiome, it is limited by low sensitivity. Enrichment using cRNA biotinylated probes has been applied to address this limitation, enhancing the detection of rare ARGs and MGEs, especially when combined with long-read sequencing. Here, we present the TELCoMB protocol, a Snakemake workflow that elucidates resistome and mobilome composition and diversity and uncovers ARG-MGE colocalizations. The protocol supports both short- and long-read sequencing and does not require enrichment, making it versatile for various genomic data types. TELCoMB generates publication-ready figures and CSV files for comprehensive analysis, improving our understanding of antimicrobial resistance mechanisms and spread. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Installing TELCOMB Locally</p><p><b>Alternate Protocol</b>: Installing TELCOMB on a SLURM Cluster</p><p><b>Basic Protocol 2</b>: Data Preprocessing</p><p><b>Basic Protocol 3</b>: Calculation of Resistome Distribution and Composition</p><p><b>Basic Protocol 4</b>: Identification of ARG-MGE Colocalizations</p>\",\"PeriodicalId\":93970,\"journal\":{\"name\":\"Current protocols\",\"volume\":\"4 10\",\"pages\":\"\"},\"PeriodicalIF\":0.0000,\"publicationDate\":\"2024-10-24\",\"publicationTypes\":\"Journal Article\",\"fieldsOfStudy\":null,\"isOpenAccess\":false,\"openAccessPdf\":\"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.70031\",\"citationCount\":\"0\",\"resultStr\":null,\"platform\":\"Semanticscholar\",\"paperid\":null,\"PeriodicalName\":\"Current protocols\",\"FirstCategoryId\":\"1085\",\"ListUrlMain\":\"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.70031\",\"RegionNum\":0,\"RegionCategory\":null,\"ArticlePicture\":[],\"TitleCN\":null,\"AbstractTextCN\":null,\"PMCID\":null,\"EPubDate\":\"\",\"PubModel\":\"\",\"JCR\":\"\",\"JCRName\":\"\",\"Score\":null,\"Total\":0}","platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.70031","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 0
The TELCoMB Protocol for High-Sensitivity Detection of ARG-MGE Colocalizations in Complex Microbial Communities
Understanding the genetic basis of antimicrobial resistance is crucial for developing effective mitigation strategies. One necessary step is to identify the antimicrobial resistance genes (ARGs) within a microbial population, referred to as the resistome, as well as the mobile genetic elements (MGEs) harboring ARGs. Although shotgun metagenomics has been successful in detecting ARGs and MGEs within a microbiome, it is limited by low sensitivity. Enrichment using cRNA biotinylated probes has been applied to address this limitation, enhancing the detection of rare ARGs and MGEs, especially when combined with long-read sequencing. Here, we present the TELCoMB protocol, a Snakemake workflow that elucidates resistome and mobilome composition and diversity and uncovers ARG-MGE colocalizations. The protocol supports both short- and long-read sequencing and does not require enrichment, making it versatile for various genomic data types. TELCoMB generates publication-ready figures and CSV files for comprehensive analysis, improving our understanding of antimicrobial resistance mechanisms and spread. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1: Installing TELCOMB Locally
Alternate Protocol: Installing TELCOMB on a SLURM Cluster
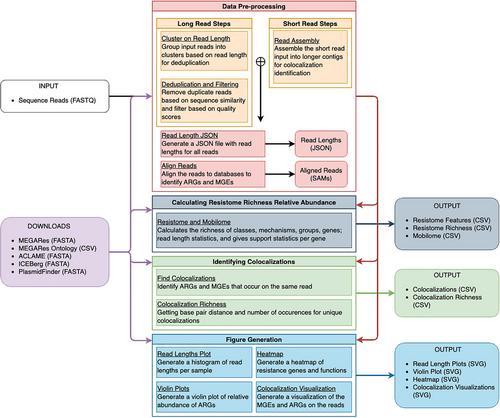
Basic Protocol 2: Data Preprocessing
Basic Protocol 3: Calculation of Resistome Distribution and Composition
Basic Protocol 4: Identification of ARG-MGE Colocalizations

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


