通过基于深度点图表示的隐式场对肺树结构进行高效解剖标记。
IF 10.7
1区 医学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
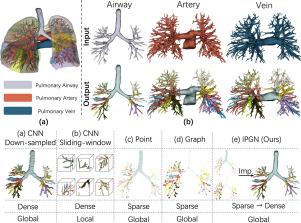
肺部疾病是导致全球死亡的主要原因之一。治疗肺病需要更好地了解肺部系统内复杂的三维树状结构,如气道、动脉和静脉等。在密集体素网格上使用高分辨率图像堆栈和标准 CNN 的传统方法在计算效率、有限分辨率、局部上下文和形状拓扑保存不足等方面面临挑战。我们的方法通过从密集体素转向稀疏点表示来解决这些问题,从而提供更好的内存效率和全局上下文利用率。然而,点表示法固有的稀疏性可能会导致树形结构中关键连接性的丢失。为了缓解这一问题,我们在骨架化结构上引入了图学习,并结合了可变特征融合,以改进拓扑结构和远距离上下文捕捉。此外,我们还采用了隐式函数,将稀疏表示高效地转换为端到端的密集重构。所提出的方法不仅在整体和关键位置的标注准确性上达到了最先进的水平,而且还能进行高效推理并生成封闭的表面形状。为了解决该领域数据稀缺的问题,我们还策划了一个综合数据集来验证我们的方法。数据和代码见 https://github.com/M3DV/pulmonary-tree-labeling。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Efficient anatomical labeling of pulmonary tree structures via deep point-graph representation-based implicit fields
Pulmonary diseases rank prominently among the principal causes of death worldwide. Curing them will require, among other things, a better understanding of the complex 3D tree-shaped structures within the pulmonary system, such as airways, arteries, and veins. Traditional approaches using high-resolution image stacks and standard CNNs on dense voxel grids face challenges in computational efficiency, limited resolution, local context, and inadequate preservation of shape topology. Our method addresses these issues by shifting from dense voxel to sparse point representation, offering better memory efficiency and global context utilization. However, the inherent sparsity in point representation can lead to a loss of crucial connectivity in tree-shaped structures. To mitigate this, we introduce graph learning on skeletonized structures, incorporating differentiable feature fusion for improved topology and long-distance context capture. Furthermore, we employ an implicit function for efficient conversion of sparse representations into dense reconstructions end-to-end. The proposed method not only delivers state-of-the-art performance in labeling accuracy, both overall and at key locations, but also enables efficient inference and the generation of closed surface shapes. Addressing data scarcity in this field, we have also curated a comprehensive dataset to validate our approach. Data and code are available at https://github.com/M3DV/pulmonary-tree-labeling.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Medical image analysis
工程技术-工程:生物医学
CiteScore
22.10
自引率
6.40%
发文量
309
审稿时长
6.6 months
期刊介绍:
Medical Image Analysis serves as a platform for sharing new research findings in the realm of medical and biological image analysis, with a focus on applications of computer vision, virtual reality, and robotics to biomedical imaging challenges. The journal prioritizes the publication of high-quality, original papers contributing to the fundamental science of processing, analyzing, and utilizing medical and biological images. It welcomes approaches utilizing biomedical image datasets across all spatial scales, from molecular/cellular imaging to tissue/organ imaging.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


