利用 Translatomer 对核糖体图谱进行深度学习预测,揭示翻译调控并解释疾病变异
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
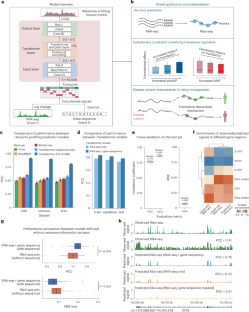
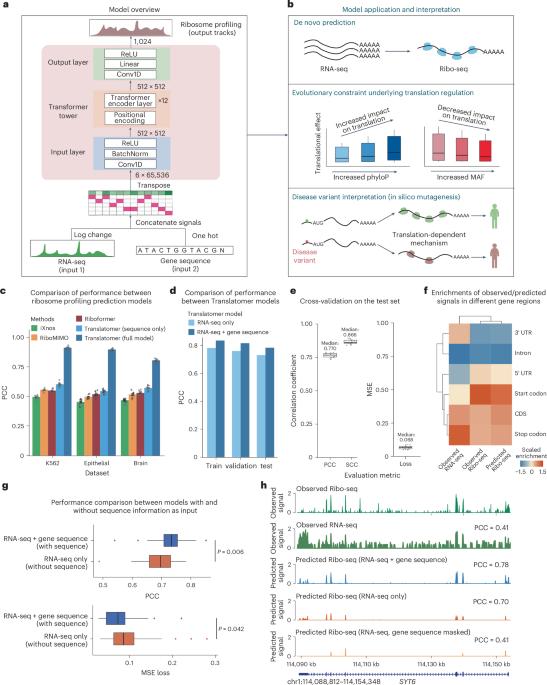
基因表达涉及转录和翻译。尽管有大量的数据集和越来越强大的方法来计算基因变异对转录的影响,但信使 RNA 和蛋白质水平之间的差异阻碍了对疾病相关变异的调控效应的系统解释。要缩小这一差距,解释作用于翻译的疾病相关变异,需要翻译序列决定因素的精确模型。我们在此介绍 Translatomer,这是一个多模式转换器框架,可从信使 RNA 表达和基因序列预测细胞类型特异性翻译。我们在 33 种组织和细胞系上对 Translatomer 进行了训练,结果表明,加入序列能改善核糖体剖析信号的预测,这表明 Translatomer 捕捉到了依赖序列的翻译调控信息。Translatomer 对细胞类型特异性核糖体图谱的从头预测准确率达到了 0.72 到 0.80。我们开发了一种硅学诱变工具来估计突变对翻译的影响,并证明与翻译调控相关的变体在人类群体和不同物种中都受到进化的限制。特别是,我们确定了细胞类型特异性翻译调控机制,这些机制独立于与阿尔茨海默病、精神分裂症和先天性心脏病等复杂疾病相关的 3,041 个非编码变异和同义变异的表达量性状位点。Translatomer 能准确模拟翻译的遗传基础,弥合信使 RNA 和蛋白质水平之间的差距,并为无法解读的疾病变异提供有价值的机理见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Deep learning prediction of ribosome profiling with Translatomer reveals translational regulation and interprets disease variants
Gene expression involves transcription and translation. Despite large datasets and increasingly powerful methods devoted to calculating genetic variants’ effects on transcription, discrepancy between messenger RNA and protein levels hinders the systematic interpretation of the regulatory effects of disease-associated variants. Accurate models of the sequence determinants of translation are needed to close this gap and to interpret disease-associated variants that act on translation. Here we present Translatomer, a multimodal transformer framework that predicts cell-type-specific translation from messenger RNA expression and gene sequence. We train the Translatomer on 33 tissues and cell lines, and show that the inclusion of sequence improves the prediction of ribosome profiling signal, indicating that the Translatomer captures sequence-dependent translational regulatory information. The Translatomer achieves accuracies of 0.72 to 0.80 for the de novo prediction of cell-type-specific ribosome profiling. We develop an in silico mutagenesis tool to estimate mutational effects on translation and demonstrate that variants associated with translation regulation are evolutionarily constrained, both in the human population and across species. In particular, we identify cell-type-specific translational regulatory mechanisms independent of the expression quantitative trait loci for 3,041 non-coding and synonymous variants associated with complex diseases, including Alzheimer’s disease, schizophrenia and congenital heart disease. The Translatomer accurately models the genetic underpinnings of translation, bridging the gap between messenger RNA and protein levels as well as providing valuable mechanistic insights for uninterpreted disease variants. A transformer-based approach called Translatomer is presented, which models cell-type-specific translation from messenger RNA expression and gene sequence, bridging the gap between messenger RNA and protein levels as well as providing a mechanistic insight into the genetic regulation of translation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


