全基因组范围的物种划分以及eriophyoid螨Epitrimerus sabinae复合体(螨形目:Eriophyoidea)引种程度的量化。
IF 3.6
1区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
物种复合体阻碍了对陆生物种多样性的探索,尤其是在形态上无法相互区分的小型节肢动物类群中。Eriophyoidea 中的 Epitrimerus sabinae 复合体为物种复合体的划分提供了一个有价值的案例研究,因为它们表现出有限的形态变异。在这项研究中,我们通过全基因组测序从 55 个 E. sabinae 复合体标本中获得了数千个核基因组单核苷酸多态性,涵盖了它们潜在的所有已知分布范围。我们建立了一个推断隐性物种的框架,其中包括通过系统发育和遗传聚类来确定潜在物种,然后通过种群人口学评估来确认种系的独立性(进而确定物种地位)。我们的研究结果表明,E. sabinae 复合体包括 10 个不同的物种。这些物种既有高度分化、基因隔离的品系,也有涉及基因流动的分化种群。这种基因流跨越物种边界的现象非常普遍,表明它们之间存在潜在的基因导入。此外,人口学分析表明,这十个物种在第四纪期间的大小变化轨迹独特。时间校准系统进化进一步表明,E. sabinae复合体的物种分化发生得很快,导致了新近纪时期的快速辐射。总之,平行/融合和涉及多基因流的近期分化可能解释了沙棘藻复合体的同源现象。我们的研究结果凸显了物种复合体划分的综合方法。本文章由计算机程序翻译,如有差异,请以英文原文为准。
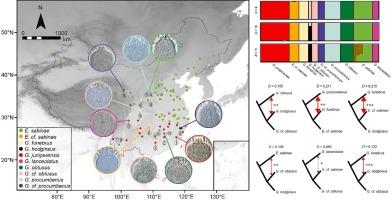
Genome-wide species delimitation and quantification of the extent of introgression in eriophyoid mite Epitrimerus sabinae complex (Acariformes: Eriophyoidea)
Species complex hinders the exploration of terrestrial species diversity, particularly in small arthropod lineages that are morphologically indistinguishable from each other. The Epitrimerus sabinae complex in the Eriophyoidea provides a valuable case study in species complex delimitation, as they exhibit limited morphological variations. In this study, we obtained thousands of nuclear genomic single-nucleotide polymorphisms via whole-genome sequencing from 55 E. sabinae complex specimens, covering their potential all known distribution ranges. We implemented a framework to infer cryptic speciation, which involved phylogenetic and genetic clustering to identify potential species, followed by population demographic assessment to confirm lineage independence (and thus species status). Our results demonstrate that the E. sabinae complex comprises ten distinct species. These species range from highly divergent, genetically isolated lineages, to differentiated populations involving gene flow. This gene flow is widespread across species boundaries, indicating potential genetic introgression among them. Additionally, demographic analyses revealed that the ten species have followed unique trajectories in size change during the Quaternary period. Time-calibrated phylogenies further showed that speciation in the E. sabinae complex occurred rapidly, resulting in a rapid radiation during the Neogene period. Collectively, parallelism/convergence and recent divergence involving multiple gene flows may explain the homoplasy of E. sabinae complex. Our results highlight the integrated approach in species complex delimitation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
7.50
自引率
7.30%
发文量
249
审稿时长
7.5 months
期刊介绍:
Molecular Phylogenetics and Evolution is dedicated to bringing Darwin''s dream within grasp - to "have fairly true genealogical trees of each great kingdom of Nature." The journal provides a forum for molecular studies that advance our understanding of phylogeny and evolution, further the development of phylogenetically more accurate taxonomic classifications, and ultimately bring a unified classification for all the ramifying lines of life. Phylogeographic studies will be considered for publication if they offer EXCEPTIONAL theoretical or empirical advances.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


