亚洲库克里蛇(Oligodon Fitzinger,1826 年)的多基因系统发育:将第二大蛇类辐射(爬行纲:有鳞目:眼镜蛇科)的刀刃磨得更锋利。
IF 3.6
1区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
乌梢蛇属(Oligodon Fitzinger)拥有 90 个公认的物种,是世界上第二大蛇类分布区。Oligodon物种集体分布于亚洲大陆,拥有其他蛇类所独有的几种生态和形态特征。尽管 Oligodon 的物种丰富度很高,但由于早期系统进化中的样本和遗传标记数量有限,人们对其内部的进化关系知之甚少。在这项研究中,我们收集了迄今为止最大的 Oligodon 分子数据集,用来评估整个蛇属的系统学和生物地理学。基于最大似然法和贝叶斯系统发育法(使用三个线粒体基因片段(12 s、16 s、CytB)和三个核基因片段(Rag1、C-mos、BDNF)),我们在 Oligodon 中发现了八个深度分化的支系,其中只有两个支系与之前形态学分类所确认的物种组相对应。对数据集线粒体部分采用的四种物种划分方法得出的分子操作分类单元(mOTUs)估计值差异巨大。综合这四种方法,我们发现了一些未被确认的种系的存在,但同时也表明其他几种 Oligodon 在遗传学上的分化程度很低,需要进行更多的综合分类研究才能正确分辨。根据分化年代测定,我们证明 Oligodon 在新近纪早期开始分化,并假设该属最近的共同祖先起源于东南亚大陆。最后,我们确认了八个系统发育明确的物种群,并指出了取样缺口,一旦有了新的数据,就需要进一步调查。这项研究有助于加深对亚洲大陆蛇类进化的了解,并为今后对这一多物种蛇属的研究奠定了基础。本文章由计算机程序翻译,如有差异,请以英文原文为准。
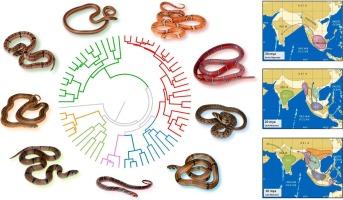
A multi-gene phylogeny of the Asian kukri snakes (Oligodon Fitzinger, 1826): Sharpening the blade of the second largest serpent radiation (Reptilia: Squamata: Colubridae)
With 90 recognized species, kukri snakes in the genus Oligodon Fitzinger constitute the second largest snake radiation in the world. Oligodon species are collectively distributed across the Asian continent and possess several ecological and morphological attributes that are unique amongst other snakes. Despite their high levels of species richness, evolutionary relationships within Oligodon are poorly understood due to a limited number of samples and genetic markers available in earlier phylogenies. In this study, we assembled the largest molecular dataset of Oligodon to date, which we use to assess the systematics and biogeography of the entire genus. Based on a combination of maximum likelihood and Bayesian phylogenies using fragments of three mitochondrial genes (12 s, 16 s, CytB) and three nuclear genes (Rag1, C-mos, BDNF), we identify eight deeply divergent clades within Oligodon, of which only two correspond with species groupings that were recognized by previous morphological classifications. Four species delimitation methods employed on the mitochondrial portion of the dataset resulted in dramatically divergent estimations of molecular operational taxonomic units (mOTUs). When combined, all four methods support the existence of unrecognized species-level lineages, but also indicate that several other Oligodon species are poorly differentiated genetically and require additional integrative taxonomic research to properly resolve. Based on divergence dating, we demonstrate that Oligodon began to diversify during the early Neogene and hypothesize that the most recent common ancestor of the genus originated in mainland Southeast Asia. We conclude by recognizing eight phylogenetically defined species groups and identify sampling gaps that require further investigation once new data becomes available. This study contributes to a greater understanding of snake evolution on the Asian continent and acts as a baseline for future studies of this speciose genus.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
7.50
自引率
7.30%
发文量
249
审稿时长
7.5 months
期刊介绍:
Molecular Phylogenetics and Evolution is dedicated to bringing Darwin''s dream within grasp - to "have fairly true genealogical trees of each great kingdom of Nature." The journal provides a forum for molecular studies that advance our understanding of phylogeny and evolution, further the development of phylogenetically more accurate taxonomic classifications, and ultimately bring a unified classification for all the ramifying lines of life. Phylogeographic studies will be considered for publication if they offer EXCEPTIONAL theoretical or empirical advances.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


