利用三维混沌博弈表示法量化 DNA 序列相似性,并将其应用于分层聚类。
IF 1.9
4区 数学
Q2 BIOLOGY
引用次数: 0
摘要
三维混沌游戏是对 DNA 序列进行编码的有效方法。由于在三维混沌游戏编码中,DNA 中的匹配子序列在空间上趋同,DNA 序列的三维混沌游戏表示法可用于比较 DNA 序列,而无需事先进行比对,也无需截断或填充任何序列。受形状相似性比较技术启发而提出的两种方法表明,这种编码方式在构建系统发生树方面与基于比对的技术一样出色。第一种方法使用相交球体的体积重叠,第二种方法通过总结三维混沌游戏轨迹的坐标、定向角和定向距离来使用形状特征。对这些方法进行了测试:(1) 11 个物种的β-球蛋白基因的第一个外显子;(2) 四组灵长类动物的线粒体 DNA;(3) 一组合成 DNA 序列。模拟结果表明,所提出的方法产生的距离能够反映突变事件的数量;此外,平均而言,缺失突变产生的距离与置换突变产生的距离相当。本文章由计算机程序翻译,如有差异,请以英文原文为准。
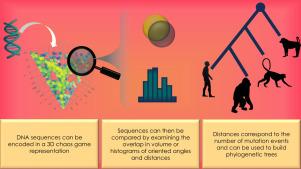
Use of 3D chaos game representation to quantify DNA sequence similarity with applications for hierarchical clustering
A 3D chaos game is shown to be a useful way for encoding DNA sequences. Since matching subsequences in DNA converge in space in 3D chaos game encoding, a DNA sequence’s 3D chaos game representation can be used to compare DNA sequences without prior alignment and without truncating or padding any of the sequences. Two proposed methods inspired by shape-similarity comparison techniques show that this form of encoding can perform as well as alignment-based techniques for building phylogenetic trees. The first method uses the volume overlap of intersecting spheres and the second uses shape signatures by summarizing the coordinates, oriented angles, and oriented distances of the 3D chaos game trajectory. The methods are tested using: (1) the first exon of the beta-globin gene for 11 species, (2) mitochondrial DNA from four groups of primates, and (3) a set of synthetic DNA sequences. Simulations show that the proposed methods produce distances that reflect the number of mutation events; additionally, on average, distances resulting from deletion mutations are comparable to those produced by substitution mutations.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
4.20
自引率
5.00%
发文量
218
审稿时长
51 days
期刊介绍:
The Journal of Theoretical Biology is the leading forum for theoretical perspectives that give insight into biological processes. It covers a very wide range of topics and is of interest to biologists in many areas of research, including:
• Brain and Neuroscience
• Cancer Growth and Treatment
• Cell Biology
• Developmental Biology
• Ecology
• Evolution
• Immunology,
• Infectious and non-infectious Diseases,
• Mathematical, Computational, Biophysical and Statistical Modeling
• Microbiology, Molecular Biology, and Biochemistry
• Networks and Complex Systems
• Physiology
• Pharmacodynamics
• Animal Behavior and Game Theory
Acceptable papers are those that bear significant importance on the biology per se being presented, and not on the mathematical analysis. Papers that include some data or experimental material bearing on theory will be considered, including those that contain comparative study, statistical data analysis, mathematical proof, computer simulations, experiments, field observations, or even philosophical arguments, which are all methods to support or reject theoretical ideas. However, there should be a concerted effort to make papers intelligible to biologists in the chosen field.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


