癌症生物标记物 microRNA-29a (miR-29a) 和 microRNA-144 (miR-144) 的混合物如何与石墨烯量子点相互作用
IF 2.7
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
据报道,微小 RNA(miRNA)是一种小型非编码 RNA,是潜在的癌症生物标志物。然而,从样本基质中提取这种短 RNA 十分困难。因此需要新的有效策略。最近,石墨烯量子点(GQDs)已在许多生物传感器平台中用于检测核苷酸,但其在 miRNA 提取方面的应用仍然有限。据报道,GQD 能够收集短 miRNA,但其收集不同结构 miRNA 的性能仍然未知。因此,这项工作研究了 GQD 与两种不同 miRNA 相互作用的能力。发夹状 miR-29a 和环状 miR-144 分子的混合物被用作两种 miRNA 形态的代表。比较研究了两个系统(miRNA 混合物,包括 4 个 miR-29a 和 4 个 miR-144,含(miR_GQD)和不含 GQD(miR))。混合物(miR)中的 miRNA 可以聚集,但没有捕获到永久性的 miRNA 集合。相比之下,GQD 的存在能迅速、自发地激活永久性 miRNA/GQD 聚集。这一发现凸显了 GQD 成为 miRNA 收集器的能力。有趣的是,所有与 GQD 结合的 miRNA 都不会展开。这使得探针很容易进入。此外,纳米级 GQD 似乎更喜欢发夹型 miR-29a。miR-29a 的自由 5' 末端是粘附在 GQD 上的粘性末端。这项工作强调了 RNA 二级结构对 GQD/miRNA 聚集能力的重要性。这一发现将有助于进一步设计 miRNA 分离策略。本文章由计算机程序翻译,如有差异,请以英文原文为准。
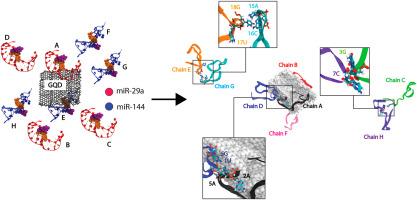
How a mixture of microRNA-29a (miR-29a) and microRNA-144 (miR-144) cancer biomarkers interacts with a graphene quantum dot
MicroRNAs (miRNAs) which are small non-coding RNAs have been reported to be potential cancer biomarker. However, it is difficult to extract such short RNA from a sample matrix. New effective strategies are required. Recently, graphene quantum dots (GQDs) have been used to detect nucleotides in many biosensor platforms, but their applications for miRNA extraction remain limited. GQD was reported to be able to collect short miRNA, but its performance to collect miRNAs with different structure remains unknown. Thus, in this work, the capability of GQD to interact with two different miRNAs is investigated. A mixture of hairpin-like miR-29a and circular miR-144 molecules are used as a representative of two miRNA morphologies. Two systems (a miRNA mixture, comprising 4 of miR-29a and 4 of miR-144, with (miR_GQD) and without GQD (miR)) were studied in comparison. MiRNAs in a mixture (miR) can aggregate, but no permanent miRNA assembly is captured. In contrast, the presence of GQD can rapidly and spontaneously activate the permanent miRNA/GQD clustering. This finding highlights the ability of GQD to be a miRNA collector. Interestingly, all GQD-bound miRNAs do not unfold. This allows the easy accessibility for probes. Also, nano-sized GQD seems to prefer hairpin miR-29a. The free 5’ terminus of miR-29a acts as the sticky end to adhere on GQD. This work highlights the importance of RNA secondary structure on GQD/miRNA aggregation capability. An insight obtained here will be useful for further design of miRNA isolation strategies.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of molecular graphics & modelling
生物-计算机:跨学科应用
CiteScore
5.50
自引率
6.90%
发文量
216
审稿时长
35 days
期刊介绍:
The Journal of Molecular Graphics and Modelling is devoted to the publication of papers on the uses of computers in theoretical investigations of molecular structure, function, interaction, and design. The scope of the journal includes all aspects of molecular modeling and computational chemistry, including, for instance, the study of molecular shape and properties, molecular simulations, protein and polymer engineering, drug design, materials design, structure-activity and structure-property relationships, database mining, and compound library design.
As a primary research journal, JMGM seeks to bring new knowledge to the attention of our readers. As such, submissions to the journal need to not only report results, but must draw conclusions and explore implications of the work presented. Authors are strongly encouraged to bear this in mind when preparing manuscripts. Routine applications of standard modelling approaches, providing only very limited new scientific insight, will not meet our criteria for publication. Reproducibility of reported calculations is an important issue. Wherever possible, we urge authors to enhance their papers with Supplementary Data, for example, in QSAR studies machine-readable versions of molecular datasets or in the development of new force-field parameters versions of the topology and force field parameter files. Routine applications of existing methods that do not lead to genuinely new insight will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


