单细胞刺激-反应基因表达轨迹揭示了单个巨噬细胞动态反应的刺激特异性
IF 14.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
巨噬细胞会诱导数百种基因的表达,以应对免疫威胁。然而,目前的技术限制了我们捕捉单细胞诱导基因表达动态的能力。在这里,我们生成了小鼠巨噬细胞对六种刺激做出反应的高分辨率时间序列单细胞 RNA 测序(scRNA-seq)数据,并对实时单细胞基因表达轨迹(scGETs)进行了估算。我们发现,scGETs 中包含的动态信息极大地促进了巨噬细胞刺激-反应特异性(SRS)。动态信息还确定了免疫反应基因之间的相关性,表明了生物协调性。此外,我们还发现极化细胞因子的微环境背景对 scGETs 有深远影响,因此与单个时间点测量相比,测量反应动态能更清晰地辨别单个巨噬细胞的极化状态。我们的研究结果凸显了免疫基因单细胞表达反应所包含的动态信息在描述巨噬细胞的SRS和功能状态方面的重要贡献。本文章由计算机程序翻译,如有差异,请以英文原文为准。
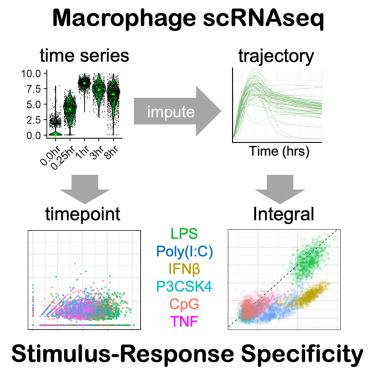
Single-cell stimulus-response gene expression trajectories reveal the stimulus specificities of dynamic responses by single macrophages
Macrophages induce the expression of hundreds of genes in response to immune threats. However, current technology limits our ability to capture single-cell inducible gene expression dynamics. Here, we generated high-resolution time series single-cell RNA sequencing (scRNA-seq) data from mouse macrophages responding to six stimuli, and imputed ensembles of real-time single-cell gene expression trajectories (scGETs). We found that dynamic information contained in scGETs substantially contributes to macrophage stimulus-response specificity (SRS). Dynamic information also identified correlations between immune response genes, indicating biological coordination. Furthermore, we showed that the microenvironmental context of polarizing cytokines profoundly affects scGETs, such that measuring response dynamics offered clearer discrimination of the polarization state of individual macrophage cells than single time-point measurements. Our findings highlight the important contribution of dynamic information contained in the single-cell expression responses of immune genes in characterizing the SRS and functional states of macrophages.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Molecular Cell
生物-生化与分子生物学
CiteScore
26.00
自引率
3.80%
发文量
389
审稿时长
1 months
期刊介绍:
Molecular Cell is a companion to Cell, the leading journal of biology and the highest-impact journal in the world. Launched in December 1997 and published monthly. Molecular Cell is dedicated to publishing cutting-edge research in molecular biology, focusing on fundamental cellular processes. The journal encompasses a wide range of topics, including DNA replication, recombination, and repair; Chromatin biology and genome organization; Transcription; RNA processing and decay; Non-coding RNA function; Translation; Protein folding, modification, and quality control; Signal transduction pathways; Cell cycle and checkpoints; Cell death; Autophagy; Metabolism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


