GGAS2SN:用于溶解度预测的门控图和 SmilesToSeq 网络
IF 5.6
2区 化学
Q1 CHEMISTRY, MEDICINAL
引用次数: 0
摘要
水溶性是药物研发中的一个关键理化特性。溶解度是药物开发中的一个关键问题,因为它可以限制药物的吸收能力。准确的溶解度预测对于药理学、环境和药物开发研究至关重要。本研究通过将门控图神经网络(GGNN)和图注意神经网络(GAT)与 Smiles2Seq 编码相结合,介绍了一种新的溶解度预测方法。我们的方法包括将化合物转换成图结构,其中节点代表原子,边表示化学键。然后使用专门的图神经网络(GNN)架构对这些图进行处理。将注意机制纳入 GNN 可以捕捉微妙的结构依赖关系,从而提高溶解度预测能力。此外,我们还利用 Smiles2Seq 编码技术弥合分子结构与其文本表述之间的语义差距。Smiles2Seq 能将化学符号无缝转换为数字序列,从而促进信息高效地传输到我们的模型中。我们通过对基准溶解度数据集的全面实验证明了我们方法的有效性,展示了与传统方法相比更优越的预测性能。我们的模型优于现有的溶解度预测模型,并对驱动溶解度行为的分子特征提供了可解释的见解。这项研究标志着溶解度预测领域的重要进步,为药物发现、制剂开发和环境评估提供了有力的工具。GGNN 和 Smiles2Seq 编码的融合为准确预测各种化合物的溶解度建立了一个强大的框架,促进了依赖溶解度数据的各个领域的创新。本文章由计算机程序翻译,如有差异,请以英文原文为准。
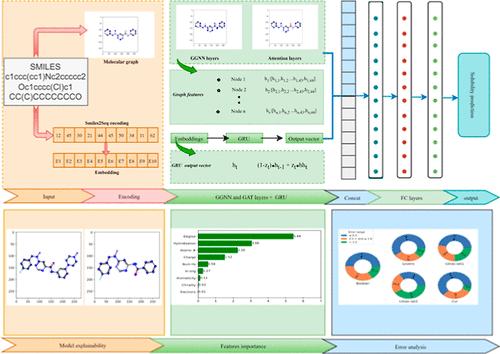
GGAS2SN: Gated Graph and SmilesToSeq Network for Solubility Prediction
Aqueous solubility is a critical physicochemical property of drug discovery. Solubility is a key issue in pharmaceutical development because it can limit a drug’s absorption capacity. Accurate solubility prediction is crucial for pharmacological, environmental, and drug development studies. This research introduces a novel method for solubility prediction by combining gated graph neural networks (GGNNs) and graph attention neural networks (GATs) with Smiles2Seq encoding. Our methodology involves converting chemical compounds into graph structures with nodes representing atoms and edges indicating chemical bonds. These graphs are then processed by using a specialized graph neural network (GNN) architecture. Incorporating attention mechanisms into GNN allows for capturing subtle structural dependencies, fostering improved solubility predictions. Furthermore, we utilized the Smiles2Seq encoding technique to bridge the semantic gap between molecular structures and their textual representations. Smiles2Seq seamlessly converts chemical notations into numeric sequences, facilitating the efficient transfer of information into our model. We demonstrate the efficacy of our approach through comprehensive experiments on benchmark solubility data sets, showcasing superior predictive performance compared to traditional methods. Our model outperforms existing solubility prediction models and provides interpretable insights into the molecular features driving solubility behavior. This research signifies an important advancement in solubility prediction, offering potent tools for drug discovery, formulation development, and environmental assessments. The fusion of GGNN and Smiles2Seq encoding establishes a robust framework for accurately forecasting solubility across various chemical compounds, fostering innovation in various domains reliant on solubility data.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
9.80
自引率
10.70%
发文量
529
审稿时长
1.4 months
期刊介绍:
The Journal of Chemical Information and Modeling publishes papers reporting new methodology and/or important applications in the fields of chemical informatics and molecular modeling. Specific topics include the representation and computer-based searching of chemical databases, molecular modeling, computer-aided molecular design of new materials, catalysts, or ligands, development of new computational methods or efficient algorithms for chemical software, and biopharmaceutical chemistry including analyses of biological activity and other issues related to drug discovery.
Astute chemists, computer scientists, and information specialists look to this monthly’s insightful research studies, programming innovations, and software reviews to keep current with advances in this integral, multidisciplinary field.
As a subscriber you’ll stay abreast of database search systems, use of graph theory in chemical problems, substructure search systems, pattern recognition and clustering, analysis of chemical and physical data, molecular modeling, graphics and natural language interfaces, bibliometric and citation analysis, and synthesis design and reactions databases.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


