鉴定谭羊肌肉发育途径中的关键 lncRNA 和 mRNA。
IF 2.2
2区 生物学
Q4 BIOCHEMISTRY & MOLECULAR BIOLOGY
Comparative Biochemistry and Physiology D-Genomics & Proteomics
Pub Date : 2024-09-30
DOI:10.1016/j.cbd.2024.101336
引用次数: 0
摘要
该研究旨在确定调控谭羊肌肉发育的长非编码RNA(lncRNA)。研究人员对绵羊1日龄和60日龄的背阔肌样本进行了RNA-seq分析,以研究肌肉发育的分子过程。结果发现,共有 5517 个 lncRNA 和 2885 个 mRNA 在 60 日龄谭羊体内有差异表达。基因本体(GO)和京都基因组百科全书(KEGG)富集分析表明,这些差异表达的lncRNA和mRNA与肌肉发育的关键通路有关,如MAPK、cAMP和钙介导的信号通路。CDKN1A、MAPK14、TGFB1、MEF2C、MYOD1和CD53等关键基因被鉴定为肌肉发育的重要参与者。研究通过RT-qPCR验证了RNA-seq结果,证实了不同表达的lncRNA和mRNA表达水平的一致性。这些研究结果表明,lncRNA-mRNA网络在调节谭羊肌肉发育方面产生了显著效果,如lncRNAs(MSTRG.12808.1/MSTRG.22662.3/MSTRG.18310.1)和mRNAs(MSTRG.10027/MSTRG.10029/MSTRG.10258/MSTRG.11011/MSTRG.10354),为今后该领域的研究奠定了基础。本文章由计算机程序翻译,如有差异,请以英文原文为准。
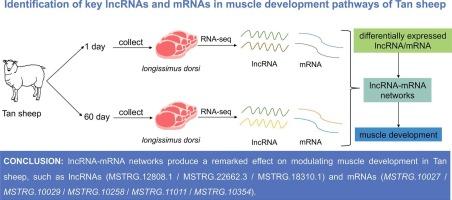
Identification of key lncRNAs and mRNAs in muscle development pathways of Tan sheep
The study aimed to identify the long noncoding RNA (lncRNA) responsible for regulating muscle development in Tan sheep. RNA-seq analysis was conducted on longissimus dorsi samples from 1-day-old and 60-day-old Tan sheep to investigate the molecular processes involved in muscle development. A total of 5517 lncRNAs and 2885 mRNAs were found to be differentially expressed in the 60-day-old Tan sheep. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis revealed that these differentially expressed lncRNAs and mRNAs were linked to pathways crucial for muscle development, such as MAPK, cAMP, and calcium-mediated signaling pathways. Key genes like CDKN1A, MAPK14, TGFB1, MEF2C, MYOD1, and CD53 were identified as significant players in muscle development. The study validated the RNA-seq results through RT-qPCR, confirming the consistency of expression levels of differentially expressed lncRNAs and mRNAs. These findings indicate that lncRNA-mRNA networks produce a remarked effect on modulating muscle development in Tan sheep, such as lncRNAs (MSTRG.12808.1/MSTRG.22662.3/MSTRG.18310.1) and mRNAs (MSTRG.10027/MSTRG.10029/MSTRG.10258/MSTRG.11011/MSTRG.10354), laying the groundwork for future research in this area.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
5.10
自引率
3.30%
发文量
69
审稿时长
33 days
期刊介绍:
Comparative Biochemistry & Physiology (CBP) publishes papers in comparative, environmental and evolutionary physiology.
Part D: Genomics and Proteomics (CBPD), focuses on “omics” approaches to physiology, including comparative and functional genomics, metagenomics, transcriptomics, proteomics, metabolomics, and lipidomics. Most studies employ “omics” and/or system biology to test specific hypotheses about molecular and biochemical mechanisms underlying physiological responses to the environment. We encourage papers that address fundamental questions in comparative physiology and biochemistry rather than studies with a focus that is purely technical, methodological or descriptive in nature.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


