Dy-SheHeRASADe:通过表面描述符表征 β 片层动力学。
IF 2.7
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
分子动力学(MD)模拟是研究原子水平大分子动态运动的重要工具。随着高性能计算能力的不断提高,分子动力学模拟的应用也越来越广泛。这使得分子建模人员能够模拟大分子结构的分子行为,获得更长的轨迹。适当的 MD 轨迹可视化对于即时直观地了解分子动力学和功能至关重要。在这项研究中,我们采用了一种新颖的三维图形表示法--表面描述符表示法的动态薄片助手(Dy-SheHeRASADe),在分子动力学背景下可视化蛋白质的β薄片二级结构。Dy-SheHeRASADe 是在开源分子浏览器和原型平台 UnityMol 中开发的。我们考虑了分子动力学模拟过程中的β片层波动和氢键形成,以描述β片层中运动较大或有易变键的部分。我们提出了两种可视化模式,它们基于根据 α 碳的位置和 β 链之间的氢键计算出来的 β 片的表面表示。体积模式:该表面被封闭在一个半透明的体积中,该体积代表薄片在动力学过程中的波动区域。热图模式:根据 α 碳的振幅值对表面进行着色。此外,我们还通过显示 β 片的最大和最小运动值、片的表面积和氢键数量来量化 β 片的波动。本文章由计算机程序翻译,如有差异,请以英文原文为准。
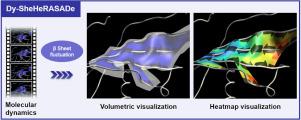
Dy-SheHeRASADe: A representation of the β sheet dynamics through surface descriptors
Molecular dynamics (MD) simulations are important tools for studying the dynamic motions of macromolecules at the atomic level. With the increasing capabilities of high performance computing, MD simulations are becoming more widely used. This allows molecular modelers to simulate the molecular behavior of large molecular architectures for much longer trajectories. Appropriate visualization of MD trajectories is becoming essential to provide an immediate and intuitive understanding of a molecule’s dynamics and function. In this study, we implement a novel 3D graphical representation, Dynamical Sheets Helper for RepresentAtion of SurfAce Descriptors (Dy-SheHeRASADe), to visualize the sheet secondary structures of proteins in the context of molecular dynamics. Dy-SheHeRASADe is developed in UnityMol, an open source molecular viewer and prototyping platform. We considered sheet fluctuations and hydrogen bond formation during molecular dynamics simulations to characterize the parts of sheets with large motions or with labile bonds. We propose two visualization modes based on a surface representation of the sheets calculated according to the positions of the carbons and the hydrogen bonds between the strands. The volumetric mode, in which this surface is enclosed in a semi-transparent volume that represents the fluctuation zone of the sheet during dynamics. The heatmap mode, in which the surface is colored according to the amplitude values of the carbons. In addition, we quantify the sheet fluctuations by displaying the values of the largest and smallest movements of the sheets, the surface area of the sheets, and the number of hydrogen bonds.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of molecular graphics & modelling
生物-计算机:跨学科应用
CiteScore
5.50
自引率
6.90%
发文量
216
审稿时长
35 days
期刊介绍:
The Journal of Molecular Graphics and Modelling is devoted to the publication of papers on the uses of computers in theoretical investigations of molecular structure, function, interaction, and design. The scope of the journal includes all aspects of molecular modeling and computational chemistry, including, for instance, the study of molecular shape and properties, molecular simulations, protein and polymer engineering, drug design, materials design, structure-activity and structure-property relationships, database mining, and compound library design.
As a primary research journal, JMGM seeks to bring new knowledge to the attention of our readers. As such, submissions to the journal need to not only report results, but must draw conclusions and explore implications of the work presented. Authors are strongly encouraged to bear this in mind when preparing manuscripts. Routine applications of standard modelling approaches, providing only very limited new scientific insight, will not meet our criteria for publication. Reproducibility of reported calculations is an important issue. Wherever possible, we urge authors to enhance their papers with Supplementary Data, for example, in QSAR studies machine-readable versions of molecular datasets or in the development of new force-field parameters versions of the topology and force field parameter files. Routine applications of existing methods that do not lead to genuinely new insight will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


