HIV-1 逆转录酶错误率和转录阈值,基于对来自体外转录细胞和 HIV 感染细胞的目标 RNA 的单链共识测序。
IF 4.7
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
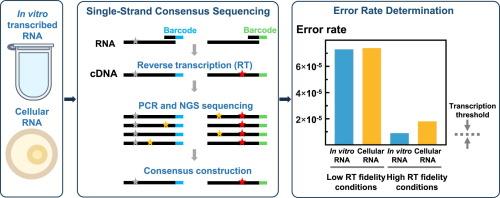
核苷酸掺入法和基于 lacZ 的正向突变检测法已被广泛用于确定反转录酶(RT)在 RNA 依赖性 DNA 聚合反应中的准确性。然而,它们涉及相当复杂和费力的程序,无法提供准确的错误率。最近,基于 NGS 的条形码方法为检测 RT 引入的所有错误提供了可能,但由于需要测序的文库规模较大,其广泛应用受到了成本的限制。在本研究中,我们介绍了一种基于单链共识测序的新颖且相对简单的 NGS 检测方法,它能在原始序列数量相对较少(约 60 Mb)的情况下提供可靠的结果。该方法以 HIV-1 蛋白酶编码序列为目标,通过测定 HIV-1(BH10 株)RT 的错误率进行了验证。在标准条件(37°C/3 mM Mg2+)下,使用体外转录的 RNA 进行 HIV-1 逆转录的错误率约为 7.3×10-5。与之前的报道一致,将 Mg2+ 浓度降至 0.5 mM 后,RT 的准确性提高了 8 倍。在 50°C 时,HIV-1 RT 的准确性也比 37°C 时高(错误率为 1.5×10-5)。有趣的是,以感染细胞中的 HIV-1 RNA 为模板,在 3 mM Mg2+(7.4×10-5)条件下进行反转录时,误差率与体外转录 RNA 测定的误差率相似,而在 0.5 mM Mg2+条件下,误差率降至 1.8×10-5。在低镁浓度下得到的数值略高于为人类细胞计算的转录错误率,从而表明我们基于 NGS 的错误率测定具有现实的转录阈值。本文章由计算机程序翻译,如有差异,请以英文原文为准。
HIV-1 Reverse Transcriptase Error Rates and Transcriptional Thresholds Based on Single-strand Consensus Sequencing of Target RNA Derived From In Vitro-transcription and HIV-infected Cells
Nucleotide incorporation and lacZ-based forward mutation assays have been widely used to determine the accuracy of reverse transcriptases (RTs) in RNA-dependent DNA polymerization reactions. However, they involve quite complex and laborious procedures, and cannot provide accurate error rates. Recently, NGS-based methods using barcodes opened the possibility of detecting all errors introduced by the RT, although their widespread use is limited by cost, due to the large size of libraries to be sequenced. In this study, we describe a novel and relatively simple NGS assay based on single-strand consensus sequencing that provides robust results with a relatively small number of raw sequences (around 60 Mb). The method has been validated by determining the error rate of HIV-1 (BH10 strain) RT using the HIV-1 protease-coding sequence as target. HIV-1 reverse transcription error rates in standard conditions (37 °C/3 mM Mg2+) using an in vitro-transcribed RNA were around 7.3 × 10−5. In agreement with previous reports, an 8-fold increase in RT’s accuracy was observed after reducing Mg2+ concentration to 0.5 mM. The fidelity of HIV-1 RT was also higher at 50 °C than at 37 °C (error rate 1.5 × 10−5). Interestingly, error rates obtained with HIV-1 RNA from infected cells as template of the reverse transcription at 3 mM Mg2+ (7.4 × 10−5) were similar to those determined with the in vitro-transcribed RNA, and were reduced to 1.8 × 10−5 in the presence of 0.5 mM Mg2+. Values obtained at low magnesium concentrations were modestly higher than the transcription error rates calculated for human cells, thereby suggesting a realistic transcriptional threshold for our NGS-based error rate determinations.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


