固态核磁共振光谱揭示 Natronomonas pharaonis halorhodopsin 在光周期后期的结构变化。
IF 3.3
3区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
Natronomonas pharaonis halorhodopsin(NpHR)是一种光驱动的Cl-内向泵,被广泛用作光遗传学工具。虽然此前对 NpHR 进行了广泛的研究,但从蛋白质结构的角度来看,对其 Cl- 摄取过程的了解并不深入,这主要是因为在晶体晶格中,很难分析与 Cl- 摄取过程相关的结构变化。在本研究中,我们利用固态核磁共振分析了 NpHR 在接近生理跨膜条件下的结合态和游离态。化学位移扰动分析表明,Cl-耗竭引起的结构变化遍及整个 NpHR 分子,但螺旋 D 细胞外(EC)部分的残基表现出显著的构象变化,这可能与 Cl-吸收过程有关。通过结合光化学分析和动态核偏振(DNP)增强固态核磁共振测量 NpHR 的点突变体,我们证实了这些残基在 Cl- 摄取过程中的重要性。特别是,我们发现位于三聚体界面的 Ala165 位氨基酸(A165V)突变为一个具有稠厚侧链的氨基酸(A165V)会显著扰乱后期的光周期,破坏其在无 Cl 状态下的三聚体组装,以及在光照射条件下的离子泵循环。这强烈表明,EC 部分的螺旋 D 向外移动,破坏了三聚体的完整性。结合光谱数据和已知的 NpHR 晶体结构,我们提出了一个模型,即这种螺旋运动是在蛋白质胞外表面创建 Cl- 入口通道所必需的,对 Cl- 吸收过程至关重要。本文章由计算机程序翻译,如有差异,请以英文原文为准。
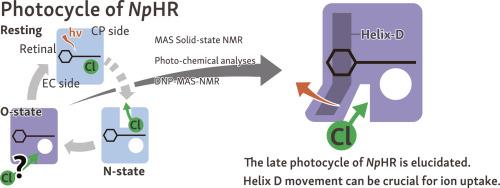
Structural changes of Natronomonas pharaonis halorhodopsin in its late photocycle revealed by solid-state NMR spectroscopy
Natronomonas pharaonis halorhodopsin (NpHR) is a light-driven Cl− inward pump that is widely used as an optogenetic tool. Although NpHR is previously extensively studied, its Cl− uptake process is not well understood from the protein structure perspective, mainly because in crystalline lattice, it has been difficult to analyze the structural changes associated with the Cl− uptake process. In this study, we used solid-state NMR to analyze NpHR both in the Cl−-bound and -free states under near-physiological transmembrane condition. Chemical shift perturbation analysis suggested that while the structural change caused by the Cl− depletion is widespread over the NpHR molecule, residues in the extracellular (EC) part of helix D exhibited significant conformational changes that may be related to the Cl− uptake process. By combining photochemical analysis and dynamic nuclear polarization (DNP)-enhanced solid-state NMR measurement on NpHR point mutants for the suggested residues, we confirmed their importance in the Cl− uptake process. In particular, we found the mutation at Ala165 position, located at the trimer interface, to an amino acid with bulky sidechain (A165V) significantly perturbs the late photocycle and disrupts its trimeric assembly in the Cl−-free state as well as during the ion-pumping cycle under the photo-irradiated condition. This strongly suggested an outward movement of helix D at EC part, disrupting the trimer integrity. Together with the spectroscopic data and known NpHR crystal structures, we proposed a model that this helix movement is required for creating the Cl− entrance path on the extracellular surface of the protein and is crucial to the Cl− uptake process.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Biophysical chemistry
生物-生化与分子生物学
CiteScore
6.10
自引率
10.50%
发文量
121
审稿时长
20 days
期刊介绍:
Biophysical Chemistry publishes original work and reviews in the areas of chemistry and physics directly impacting biological phenomena. Quantitative analysis of the properties of biological macromolecules, biologically active molecules, macromolecular assemblies and cell components in terms of kinetics, thermodynamics, spatio-temporal organization, NMR and X-ray structural biology, as well as single-molecule detection represent a major focus of the journal. Theoretical and computational treatments of biomacromolecular systems, macromolecular interactions, regulatory control and systems biology are also of interest to the journal.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


