Schizoparmaceae 科及其相关科(Diaporthales, Ascomycota)的分子系统发育和进化分异及生物地理学估计。
IF 3.6
1区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
Diaporthales 包括 32 个科,其中许多科都是重要的植物病原体、内生菌和酵母菌,如假柄菌科、锈菌科和五味子科的成员。核苷酸序列来自五个基因位点,包括ITS, LSU, TEF1-α, TUB2 和 RPB2 等五个基因位点的核苷酸序列被用于贝叶斯进化分析,以确定五味子科的分化时间和进化关系。分子钟分析表明,五味子科的祖先分裂于上白垩纪,约75.7百万年(95%最高后验密度为60.3-91.3百万年)。利用贝叶斯二元马尔科夫链蒙特卡洛法(BBM)重建系统发育中的祖先状态(RASP),重建了五味子科的历史生物地理学,表明其最有可能起源于非洲。根据分类学和系统发生学分析,明确了假柄孢科(Pseudoplagiostomataceae)和糙叶孢科(Pyrisporaceae)的关系,共描述了 4 个物种。在假柄孢科方面,描述了三个新种和一个已知种,包括 Ps.关于拟盘孢科,我们认为 Pseudoplagiostoma castaneae 是 Pyrispora castaneae 的异名。此外,我们还描述了 Schizoparmaceae 的一个新种 Coniella fujianensis sp.本文章由计算机程序翻译,如有差异,请以英文原文为准。
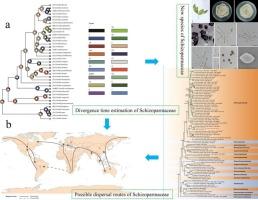
Molecular phylogenetic and estimation of evolutionary divergence and biogeography of the family Schizoparmaceae and allied families (Diaporthales, Ascomycota)
The Diaporthales includes 32 families, many of which are important plant pathogens, endophytes and saprobes, e.g., members of the families Pseudoplagiostomataceae, Pyrisporaceae and Schizoparmaceae. Nucleotide sequences derived from five genetic loci including: ITS, LSU, TEF1-α, TUB2 and RPB2 were used for Bayesian evolutionary analysis to determine divergence times and evolutionary relationships within the Schizoparmaceae. Molecular clock analyses revealed that the ancestor of Schizoparmaceae split during the Upper Cretaceous period approximately 75.7 Mya (95 % highest posterior density of 60.3–91.3 Mya). Reconstructing ancestral state in phylogenies (RASP) with using the Bayesian Binary Markov chain Monte Carlo (BBM) Method to reconstruct the historical biogeography for the family Schizoparmaceae indicated its most likely origin in Africa. Based on taxonomic and phylogenetic analyses, the Pseudoplagiostomataceae and Pyrisporaceae relationships were clarified and a total of four species described herein. For Pseudoplagiostomataceae, three new species and one known species that include, Pseudoplagiostoma fafuense sp. nov., Ps. ilicis sp. nov., Ps. sanmingense sp. nov. and Ps. bambusae are described and a key of Pseudoplagiostomataceae is provided. With respect to Pyrisporaceae, we considered Pseudoplagiostoma castaneae to be a synonym of Pyrispora castaneae. In addition, a new species of Schizoparmaceae, Coniella fujianensis sp. nov. is described and illustrated.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
7.50
自引率
7.30%
发文量
249
审稿时长
7.5 months
期刊介绍:
Molecular Phylogenetics and Evolution is dedicated to bringing Darwin''s dream within grasp - to "have fairly true genealogical trees of each great kingdom of Nature." The journal provides a forum for molecular studies that advance our understanding of phylogeny and evolution, further the development of phylogenetically more accurate taxonomic classifications, and ultimately bring a unified classification for all the ramifying lines of life. Phylogeographic studies will be considered for publication if they offer EXCEPTIONAL theoretical or empirical advances.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


