转录组数据支持系统发育的一致性,并揭示了与鲤形目鳉鱼一年生反复进化相关的基因组变化。
IF 3.6
1区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
与生态变量相关的新生活史的重复进化为研究遗传、发育和代谢特征的趋同提供了机会。鳉鱼(Aplocheiloid killifishes)是分布在环热带地区的跃层鱼类的一个支系,在其 800 个物种中,有近一半是 "年生 "或季节性物种,它们生存在短暂的水体中,这些水体每年都会干涸数周至数月,无法生长、繁殖或生存。但是,如果没有一个强大的系统发育框架,人们对这些鱼类一年生活史的关键特征--适应性的反复进化仍然知之甚少。我们利用从代表年生生活的可能独立起源的不同杀鱼系中获得的新测序的转录组,提出了一个大规模的无尾杀鱼进化系统发生组重建。对始祖状态的估计表明,发育休眠(休眠期)是鳉鱼一年生活周期中的一个关键特征,可能在非洲和南美洲的鱼系中独立起源了多达七次。为了进一步探索这一独特性状的遗传基础,我们通过使用 dN/dS 比值量化密码子进化来测量整个鳉鱼生命树中同源基因之间进化率的变化。我们发现,在导致年生活史物种的世系中,一些基因的 dN/dS 比值较高。其中许多基因构成了关键的发育基因或控制氧化磷酸化的核编码代谢基因。最后,我们将这些ω较高的基因与之前在鳉鱼和其他脊椎动物中与发育休眠和代谢转变相关的基因进行了比较,从而确定了重复过渡到极端环境的分子进化特征。本文章由计算机程序翻译,如有差异,请以英文原文为准。
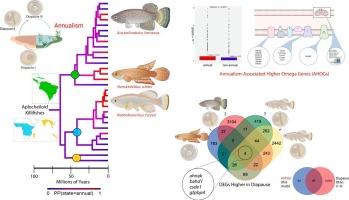
Transcriptomic data support phylogenetic congruence and reveal genomic changes associated with the repeated evolution of annualism in aplocheiloid killifishes (Cyprinodontiformes)
Repeated evolution of novel life histories that are correlated with ecological variables offers opportunities to study convergence in genetic, developmental, and metabolic features. Nearly half of the 800 species of Aplocheiloid killifishes, a clade of teleost fishes with a circumtropical distribution, are “annual” or seasonal species that survive in ephemeral bodies of water that desiccate and are unfeasible for growth, reproduction, or survival for weeks to months every year. But the repeated evolution of adaptations that are key features of the annual life history among these fishes remains poorly known without a robust phylogenetic framework. We present a large-scale phylogenomic reconstruction of aplocheiloid killifishes evolution using newly sequenced transcriptomes obtained from a diversity of killifish lineages representing putative independent origins of annualism. Ancestral state estimation shows that developmental dormancy (diapause), a key trait of the killifish annual life cycle, may have originated up to seven times independently among African and South American lineages. To further explore the genetic basis of this unique trait, we measure changes in evolutionary rates among orthologous genes across the killifish tree of life by quantifying codon evolution using dN/dS ratios. We show that some genes have higher dN/dS ratios in lineages leading to species with annual life history. Many of them constitute key developmental genes or nuclear-encoded metabolic genes that control oxidative phosphorylation. Lastly, we compare these genes with higher ω to genes previously associated to developmental dormancy and metabolic shifts in killifishes and other vertebrates, and thereby identify molecular evolutionary signatures of repeated transitions to extreme environments.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
7.50
自引率
7.30%
发文量
249
审稿时长
7.5 months
期刊介绍:
Molecular Phylogenetics and Evolution is dedicated to bringing Darwin''s dream within grasp - to "have fairly true genealogical trees of each great kingdom of Nature." The journal provides a forum for molecular studies that advance our understanding of phylogeny and evolution, further the development of phylogenetically more accurate taxonomic classifications, and ultimately bring a unified classification for all the ramifying lines of life. Phylogeographic studies will be considered for publication if they offer EXCEPTIONAL theoretical or empirical advances.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


