评估 AlphaFold3 与人类脂肪酸结合蛋白的脂肪酸对接。
IF 2.7
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
人类脂肪酸结合蛋白(FABPs)参与脂质代谢的许多方面,如亲脂性分子的摄取、运输和储存以及细胞功能。了解 FABP 如何识别脂肪酸(FA)对于确定 FABP 的功能和应用(如抑制剂设计或生物标记物开发)至关重要。最近开发的 AlphaFold3(AF3)显示出明显高于其他预测工具的准确性,特别是在预测与最先进对接工具的蛋白质配体相互作用方面。目前还缺乏有关 AF3 是否可用于识别 FABP 的 FA 的研究。为了评估使用 AF3 将 FA 与 FABP 对接的准确性,将使用 AF3 生成的 FA 与 FABP 对接的模型与实验测定的 FA 结合 FABP 结构进行了比较。AF3结构中的FA配体可靠地对接了FABPs的FA结合口袋;然而,大多数对接FABPs的FA配体的详细结合构型以及使用AF3确定的FA与FABP之间的相互作用与实验结果不同。这些结果将有助于理解使用 AF3 将 FA 与 FABPs 及其他 FA 结合蛋白对接的过程。本文章由计算机程序翻译,如有差异,请以英文原文为准。
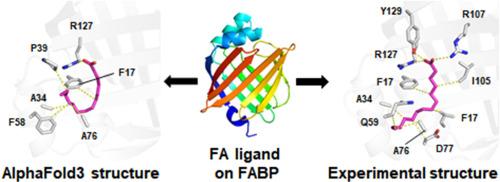
Evaluation of AlphaFold3 for the fatty acids docking to human fatty acid-binding proteins
Human fatty acid-binding proteins (FABPs) are involved in many aspects of lipid metabolism, such as the uptake, transport, and storage of lipophilic molecules, as well as cellular functions. Understanding how FABPs recognize fatty acids (FAs) is crucial for identifying FABP function and applications, such as in inhibitor design or biomarker development. The recently developed AlphaFold3 (AF3) demonstrates significantly higher accuracy than other prediction tools, particularly in predicting protein–ligand interactions with state-of-the-art docking tools. Studies on whether AF3 can be used to identify the FAs of FABP are lacking. To assess the accuracy of FA docking to FABPs using AF3, models of FA docked into FABP generated using AF3 were compared with experimentally determined FA-bound FABP structures. FA ligands in AF3 structures docked reliably into the FA-binding pocket of FABPs; however, the detailed binding configuration of most FA ligands docked into FABPs and the interaction between FA and FABP determined using AF3 and experimentally differed. These results will aid in understanding FA docking to FABPs and other FA-binding proteins using AF3.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of molecular graphics & modelling
生物-计算机:跨学科应用
CiteScore
5.50
自引率
6.90%
发文量
216
审稿时长
35 days
期刊介绍:
The Journal of Molecular Graphics and Modelling is devoted to the publication of papers on the uses of computers in theoretical investigations of molecular structure, function, interaction, and design. The scope of the journal includes all aspects of molecular modeling and computational chemistry, including, for instance, the study of molecular shape and properties, molecular simulations, protein and polymer engineering, drug design, materials design, structure-activity and structure-property relationships, database mining, and compound library design.
As a primary research journal, JMGM seeks to bring new knowledge to the attention of our readers. As such, submissions to the journal need to not only report results, but must draw conclusions and explore implications of the work presented. Authors are strongly encouraged to bear this in mind when preparing manuscripts. Routine applications of standard modelling approaches, providing only very limited new scientific insight, will not meet our criteria for publication. Reproducibility of reported calculations is an important issue. Wherever possible, we urge authors to enhance their papers with Supplementary Data, for example, in QSAR studies machine-readable versions of molecular datasets or in the development of new force-field parameters versions of the topology and force field parameter files. Routine applications of existing methods that do not lead to genuinely new insight will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


