用于膜蛋白结构研究的表达筛选和样本评估的最新进展。
IF 4.7
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
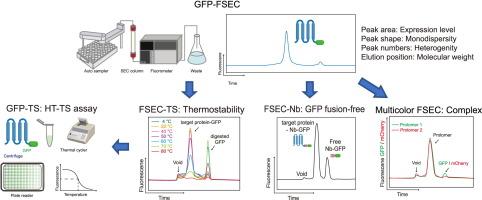
膜蛋白参与许多生物过程,占所有药物靶点的一半以上;因此,这些蛋白的结构信息非常宝贵。然而,膜蛋白表达水平低,在溶液中稳定性差,容易沉淀和聚集,是制备纯化膜蛋白用于结构研究的主要瓶颈。传统上,用于结构研究的膜蛋白构建物的评估相当耗时和昂贵,因为必须大规模表达和纯化蛋白质,特别是用于 X 射线晶体学。荧光检测尺寸排阻色谱法(FSEC)的出现彻底改变了这一局面,因为这种方法可用于快速评估小规模膜蛋白的表达和行为,而无需纯化。FSEC 已成为膜蛋白表达条件筛选和样品评估最广泛使用的方法,并成功确定了许多结构。即使在冷冻电镜时代,FSEC 和新一代 FSEC 衍生方法也以各种方式被广泛应用于结构分析。此外,FSEC 的应用不仅限于结构分析,它还广泛用于膜蛋白的功能分析,包括寡聚状态分析、抗体和配体筛选以及亲和性分析。本综述介绍了膜蛋白表达筛选和样品评估的最新进展和应用,尤其侧重于 FSEC 方法。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Recent Advances in Expression Screening and Sample Evaluation for Structural Studies of Membrane Proteins
Membrane proteins are involved in numerous biological processes and represent more than half of all drug targets; thus, structural information on these proteins is invaluable. However, the low expression level of membrane proteins, as well as their poor stability in solution and tendency to precipitate and aggregate, are major bottlenecks in the preparation of purified membrane proteins for structural studies. Traditionally, the evaluation of membrane protein constructs for structural studies has been quite time consuming and expensive since it is necessary to express and purify the proteins on a large scale, particularly for X-ray crystallography. The emergence of fluorescence detection size exclusion chromatography (FSEC) has drastically changed this situation, as this method can be used to rapidly evaluate the expression and behavior of membrane proteins on a small scale without the need for purification. FSEC has become the most widely used method for the screening of expression conditions and sample evaluation for membrane proteins, leading to the successful determination of numerous structures. Even in the era of cryo-EM, FSEC and the new generation of FSEC derivative methods are being widely used in various manners to facilitate structural analysis. In addition, the application of FSEC is not limited to structural analysis; this method is also widely used for functional analysis of membrane proteins, including for analysis of oligomerization state, screening of antibodies and ligands, and affinity profiling. This review presents the latest advances and applications in membrane protein expression screening and sample evaluation, with a particular focus on FSEC methods.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


