iACP-DFSRA:基于 ResCNN 和 Attention 双通道融合策略的抗癌肽鉴定。
IF 4.7
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
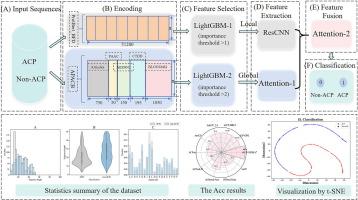
抗癌肽(ACPs)因其安全性好、副作用小、选择性高而被广泛应用于癌症治疗。然而,由于 ACPs 的鉴定费用极其昂贵,经过实验验证的 ACPs 数量有限。因此,亟需准确且经济有效的 ACPs 识别方法。在这项工作中,我们提出了一种基于深度学习的 ACPs 识别模型,命名为 iACP-DFSRA。具体来说,我们采用了两种序列嵌入技术:ProtBert_BFD 预训练语言模型和手工特征来编码蛋白质序列。然后,利用 LightGBM 进行特征选择,并将所选特征分别输入 ResCNN 和 Attention 机制,以提取局部特征和全局特征。最后,利用注意力机制对并集的特征进行深度融合,使模型更加关注关键特征,并通过全连接层进行预测。10 倍交叉验证结果表明,与最新的 AACFlow 模型相比,iACP-DFSRA 模型在大多数指标上都有所改进,Sp 为 94.15%,Sn 为 95.32%,Acc 为 94.74%,MCC 为 89.48%。事实上,iACP-DFSRA 模型是该独立测试数据集上唯一一个 Acc > 90% 和 MCC > 80% 的模型。此外,我们还在其他数据集上进一步证明了我们模型的优越性。此外,t-SNE 和 SHAP 解释分析表明,使用双通道进行特征提取和使用 Attention 机制进行深度融合至关重要,这有助于 iACP-DFSRA 更有效地预测 ACP。本文章由计算机程序翻译,如有差异,请以英文原文为准。
iACP-DFSRA: Identification of Anticancer Peptides Based on a Dual-channel Fusion Strategy of ResCNN and Attention
Anticancer peptides (ACPs) have been widely applied in the treatment of cancer owing to good safety, rational side effects, and high selectivity. However, the number of ACPs that have been experimentally validated is limited as identification of ACPs is extremely expensive. Hence, accurate and cost-effective identification methods for ACPs are urgently needed. In this work, we proposed a deep learning-based model, named iACP-DFSRA, for ACPs identification. Specifically, we adopted two kinds of sequence embedding technologies, ProtBert_BFD pre-training language model and handcrafted features to encode protein sequences. Then, the LightGBM was used for feature selection, and the selected features were input into ResCNN and Attention mechanism, respectively, to extract local and global features. Finally, the concatenate features were deeply fused by using the Attention mechanism to allow key features to be paid more attention to by the model and make predictions by fully connected layer. The results of 10-fold cross-validation demonstrated that the iACP-DFSRA model delivered improved results in most metrics with Sp of 94.15%, Sn of 95.32%, Acc of 94.74% and MCC of 89.48% compared to the latest AACFlow model. Indeed, the iACP-DFSRA model is the only model with Acc > 90% and MCC > 80% on this independent test dataset. Furthermore, we have further demonstrated the superiority of our model on additional datasets. In addition, t-SNE and SHAP interpretation analysis demonstrated that it is crucial to use two channels for feature extraction and use the Attention mechanism for deep fusion, which helps the iACP-DFSRA to predict ACPs more effectively.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


