BEMM-GEN:为分子动力学模拟生成生物分子环境模拟模型的工具包
IF 5.3
2区 化学
Q1 CHEMISTRY, MEDICINAL
引用次数: 0
摘要
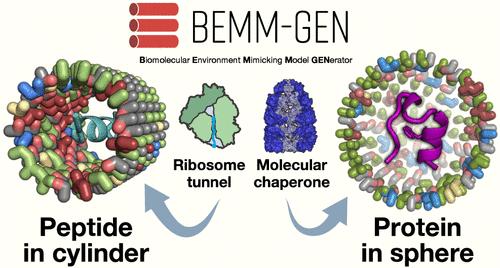
了解细胞环境对蛋白质构象的影响对于阐明活细胞内蛋白质的功能至关重要。在分子动力学(MD)模拟研究中,碳纳米管和疏水笼被广泛用于模拟特定大型生物大分子(如核糖体隧道和伴侣)内部的细胞环境。然而,最近的研究表明,这些均匀的疏水模型可能无法充分捕捉每个生物大分子内部的环境效应。基于这些事实,有必要生成与目标生物大分子内的成分相对应的具有不同化学特性的球形和圆柱形模型。我们开发了一种生物分子环境模拟模型生成器(BEMM-GEN),可生成具有用户指定化学特性的球形和圆柱形模型,并可将任意蛋白质构象整合到生成的模型中。BEMM-GEN 提供模型和蛋白质复合物结构,以及用于 MD 仿真(AMBER 和 GROMACS)的相应参数文件,用户可根据生成的输入文件立即运行 MD 仿真。BEMM-GEN 可通过 Python 软件包管理器(pip install BEMM-gen)免费下载和安装。GitHub (https://github.com/y4suda/BEMM-GEN) 上提供了源代码文件和用户操作手册。本文章由计算机程序翻译,如有差异,请以英文原文为准。
BEMM-GEN: A Toolkit for Generating a Biomolecular Environment-Mimicking Model for Molecular Dynamics Simulation
Understanding the influence of the cellular environment on protein conformations is crucial for elucidating protein functions within living cells. In studies using molecular dynamics (MD) simulation, carbon nanotubes and hydrophobic cages have been widely used to emulate the cellular environment inside specific large biomolecules such as ribosome tunnels and chaperones. However, recent studies suggest that these uniform hydrophobic models may not adequately capture the environmental effects inside each biomolecule. Based on these facts, it is necessary to generate spherical and cylindrical models with varied chemical properties corresponding to the components within target biomolecules. We developed a biomolecular environment-mimicking model generator (BEMM-GEN) that generates spherical and cylindrical models with user-specified chemical properties and allows the integration of arbitrary protein conformations into the generated models. BEMM-GEN provides model and protein complex structures, along with the corresponding parameter files for MD simulation (AMBER and GROMACS), and users immediately run their MD simulation based on the generated input files. BEMM-GEN can be freely downloaded and installed via a Python package manager (pip install BEMM-gen). The source code files and a user manual for operation are provided on GitHub (https://github.com/y4suda/BEMM-GEN).
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
9.80
自引率
10.70%
发文量
529
审稿时长
1.4 months
期刊介绍:
The Journal of Chemical Information and Modeling publishes papers reporting new methodology and/or important applications in the fields of chemical informatics and molecular modeling. Specific topics include the representation and computer-based searching of chemical databases, molecular modeling, computer-aided molecular design of new materials, catalysts, or ligands, development of new computational methods or efficient algorithms for chemical software, and biopharmaceutical chemistry including analyses of biological activity and other issues related to drug discovery.
Astute chemists, computer scientists, and information specialists look to this monthly’s insightful research studies, programming innovations, and software reviews to keep current with advances in this integral, multidisciplinary field.
As a subscriber you’ll stay abreast of database search systems, use of graph theory in chemical problems, substructure search systems, pattern recognition and clustering, analysis of chemical and physical data, molecular modeling, graphics and natural language interfaces, bibliometric and citation analysis, and synthesis design and reactions databases.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


