哺乳动物丝胶酸酯酶的结构分析。
IF 4.7
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
Sialic acid esterase(SIAE)能催化细胞表面糖蛋白中的 Sialic acids 上的 O-acetyl 基团脱落,从而调节 B 细胞受体信号传导和细胞凋亡等细胞过程。SIAE 的功能缺失突变与几种常见的自身免疫性疾病有关,包括克罗恩病、溃疡性结肠炎和关节炎。为了更好地了解这种蛋白质的功能和调控,我们测定了三种哺乳动物同源物的 SIAE 晶体结构,包括醋酸盐结合结构。这些结构显示,催化结构域采用了 SGNH水解酶超家族的折叠结构。活性位点由催化二联体组成,而不是之前报道的催化三联体。在尝试确定底物结合结构时,发现活性位点中只有水解产物醋酸盐。对完整底物的刚性对接以及随后的分子动力学模拟显示,活性位点并不与底物形成特定的相互作用,相反,它似乎具有广泛的特异性,可以接受具有不同修饰的硅聚糖。根据醋酸盐结合结构,提出了一种催化机制。疾病突变的结构图显示,大多数突变位于酶的表面,只会对蛋白质折叠造成轻微破坏,这表明这些突变可能会影响与其他因子的结合。这些结果增进了我们对 SIAE 生物学的了解,可能有助于开发治疗自身免疫性疾病和癌症的疗法。本文章由计算机程序翻译,如有差异,请以英文原文为准。
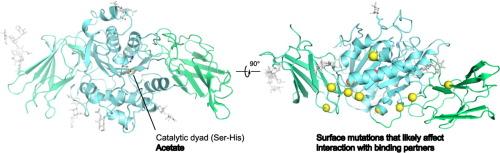
Structural Analysis of Mammalian Sialic Acid Esterase
Sialic acid esterase (SIAE) catalyzes the removal of O-acetyl groups from sialic acids found on cell surface glycoproteins to regulate cellular processes such as B cell receptor signalling and apoptosis. Loss-of-function mutations in SIAE are associated with several common autoimmune diseases including Crohn’s, ulcerative colitis, and arthritis. To gain a better understanding of the function and regulation of this protein, we determined crystal structures of SIAE from three mammalian homologs, including an acetate bound structure. The structures reveal that the catalytic domain adopts the fold of the SGNH hydrolase superfamily. The active site is composed of a catalytic dyad, as opposed to the previously reported catalytic triad. Attempts to determine a substrate-bound structure yielded only the hydrolyzed product acetate in the active site. Rigid docking of complete substrates followed by molecular dynamics simulations revealed that the active site does not form specific interactions with substrates, rather it appears to be broadly specific to accept sialoglycans with diverse modifications. Based on the acetate bound structure, a catalytic mechanism is proposed. Structural mapping of disease mutations reveals that most are located on the surface of the enzyme and would only cause minor disruptions to the protein fold, suggesting that these mutations likely affect binding to other factors. These results improve our understanding of SIAE biology and may aid in the development of therapies for autoimmune diseases and cancer.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


