合理设计非翻译区,提高基因表达。
IF 4.7
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
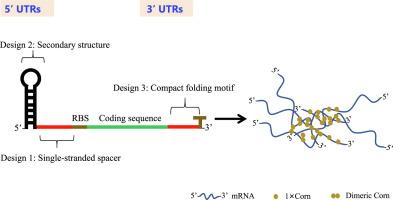
如何通过优化 mRNA 结构来改善基因表达是各种医学和生物技术应用的关键问题。以往的研究主要集中在 5' UTR 发夹结构的研究上。在本研究中,我们提出了一种合理的策略,通过对 5' 和 3' UTR 序列进行工程设计来提高 mRNA 的稳定性和翻译能力。我们以绿色荧光蛋白(GFP)为模型,在大肠杆菌和不同的表达载体中成功验证了这一策略。我们还用荧光素酶和恶性疟原虫乳酸脱氢酶(PfLDH)进一步验证了这一策略。为了阐明其基本机制,我们对蛋白质、mRNA 水平和半衰期时间进行了定量分析。我们确定了 UTR 的几个关键方面,它们对我们系统中的 mRNA 稳定性和蛋白质表达有重大影响:(1)5' UTR 中稳定发夹和核糖体结合位点(RBS)之间的单链间隔的最佳长度为 25-30 个核苷酸(nt)。间隔中 32% 的最佳 GC 含量可产生最高水平的 GFP 蛋白。(2)在 3' UTR 中插入一个可同源二聚体化的、含有 G-四重结构的 RNA 类似物 "玉米",可显著提高蛋白质的表达量。我们的研究结果表明,精心设计的 5' UTR 和 3' UTR 显著提高了基因的表达。具体来说,在 3' UTR 中加入 5×Corn 似乎能促进 mRNA 的局部聚集,从而形成 mRNA 凝聚体。除了揭示 mRNA 稳定性和表达的调控外,这项研究还有望大幅提高生物蛋白质的产量。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Rational Design of Untranslated Regions to Enhance Gene Expression
How to improve gene expression by optimizing mRNA structures is a crucial question for various medical and biotechnological applications. Previous efforts focus largely on investigation of the 5′ UTR hairpin structures. In this study, we present a rational strategy that enhances mRNA stability and translation by engineering both the 5′ and 3′ UTR sequences. We have successfully demonstrated this strategy using green fluorescent protein (GFP) as a model in Escherichia coli and across different expression vectors. We further validated it with luciferase and Plasmodium falciparum lactate dehydrogenase (PfLDH). To elucidate the underlying mechanism, we have quantitatively analyzed both protein, mRNA levels and half-life time. We have identified several key aspects of UTRs that significantly influence mRNA stability and protein expression in our system: (1) The optimal length of the single-stranded spacer between the stabilizer hairpin and ribosome binding site (RBS) in the 5′ UTR is 25–30 nucleotide (nt) long. An optimal 32% GC content in the spacer yielded the highest levels of GFP protein production. (2) The insertion of a homodimerdizable, G-quadruplex structure containing RNA aptamer, “Corn”, in the 3′ UTR markedly increased the protein expression. Our findings indicated that the carefully engineered 5′ UTRs and 3′ UTRs significantly boosted gene expression. Specifically, the inclusion of 5 × Corn in the 3′ UTR appeared to facilitate the local aggregation of mRNA, leading to the formation of mRNA condensates. Aside from shedding light on the regulation of mRNA stability and expression, this study is expected to substantially increase biological protein production.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


