核受体域间通讯由具有配体特异性的铰链介导。
IF 4.7
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
核受体是配体诱导的转录因子,可直接与目标基因结合并调节其表达。配体结合会引发构象变化,并传播到其他结构域,从而对其活性进行异构调节。人们对核受体中这种结构域间通信的性质知之甚少,这主要是由于难以通过实验鉴定全长结构。我们采用计算建模方法描述并研究了全长法尼类固醇 X 受体(FXR)的结构,该结构近似于 DNA 结合结构域(DBD)和配体结合结构域(LBD),两者通过柔性铰链区相连。利用扩展分子动力学模拟(> 10 微秒)和增强采样模拟,我们提供了配体选择性诱导结构域重排从而导致结构域间接触的证据。我们利用蛋白质-蛋白质相互作用实验提供了这些相互作用的实验证据,确定了铰链在介导结构域间接触中的关键作用。我们的研究结果揭示了 FXR 域间通讯以前未知的方面,并为其他全长核受体的特征描述提供了一个框架。本文章由计算机程序翻译,如有差异,请以英文原文为准。
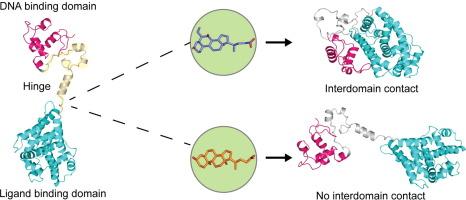
Nuclear Receptor Interdomain Communication is Mediated by the Hinge with Ligand Specificity
Nuclear receptors are ligand-induced transcription factors that bind directly to target genes and regulate their expression. Ligand binding initiates conformational changes that propagate to other domains, allosterically regulating their activity. The nature of this interdomain communication in nuclear receptors is poorly understood, largely owing to the difficulty of experimentally characterizing full-length structures. We have applied computational modeling approaches to describe and study the structure of the full-length farnesoid X receptor (FXR), approximated by the DNA binding domain (DBD) and ligand binding domain (LBD) connected by the flexible hinge region. Using extended molecular dynamics simulations (>10 microseconds) and enhanced sampling simulations, we provide evidence that ligands selectively induce domain rearrangement, leading to interdomain contact. We use protein–protein interaction assays to provide experimental evidence of these interactions, identifying a critical role of the hinge in mediating interdomain contact. Our results illuminate previously unknown aspects of interdomain communication in FXR and provide a framework to enable characterization of other full-length nuclear receptors.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


