细胞色素 c 过氧化物酶的核心、活性位点和外围区域对极端压力和温度的不同反应。
IF 4.7
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
考虑到极端环境中的生命,静水压力在原子水平上对蛋白质的影响引起了人们的极大兴趣。与环境条件相比,温度和压力的巨大偏差会改变蛋白质的自由能景观,从而揭示出原本低密度的结构状态,甚至促进蛋白质的解折。我们报告了含血红素过氧化物酶、细胞色素 c 过氧化物酶(CcP)在 1.5 和 3.0 千巴条件下的晶体结构,并将其与在 1.0 巴和低温(100 K)条件下测定的结构进行了比较。压力会使 CcP 发生各向异性的变化,但在 1.5 千巴之后可压缩性会趋于稳定。在 B 因子相对较高的蛋白质外围,CcP 对压力的反应是体积下降,但几乎保持不变的核心结构、氢键相互作用和活性位点通道。活性位点溶解和血红素连接的变化揭示了压力对蛋白质-配体相互作用和底物过氧化物潜在对接位点的敏感性。表面压缩既不会影响交替侧链构象,也不会影响 B 因子。因此,类似于结晶固体的核心填料限制了运动并保护了活性位点,而表面的松散填料则保持了侧链的动态变化。这些数据表明,蛋白质中的构象动力学和堆积密度并不完全相关,多肽对辅因子的包裹可以提供一个结构精确的环境,使其在各种物理条件下都不会发生变化。本文章由计算机程序翻译,如有差异,请以英文原文为准。
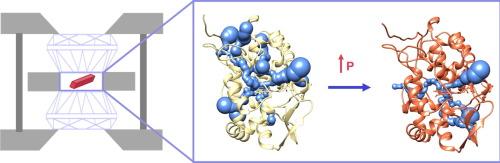
Differential Responses in the Core, Active Site and Peripheral Regions of Cytochrome c Peroxidase to Extreme Pressure and Temperature
In consideration of life in extreme environments, the effects of hydrostatic pressure on proteins at the atomic level have drawn substantial interest. Large deviations of temperature and pressure from ambient conditions can shift the free energy landscape of proteins to reveal otherwise lowly populated structural states and even promote unfolding. We report the crystal structure of the heme-containing peroxidase, cytochrome c peroxidase (CcP) at 1.5 and 3.0 kbar and make comparisons to structures determined at 1.0 bar and cryo-temperatures (100 K). Pressure produces anisotropic changes in CcP, but compressibility plateaus after 1.5 kbar. CcP responds to pressure with volume declines at the periphery of the protein where B-factors are relatively high but maintains nearly intransient core structure, hydrogen bonding interactions and active site channels. Changes in active-site solvation and heme ligation reveal pressure sensitivity to protein–ligand interactions and a potential docking site for the substrate peroxide. Compression at the surface affects neither alternate side-chain conformers nor B-factors. Thus, packing in the core, which resembles a crystalline solid, limits motion and protects the active site, whereas looser packing at the surface preserves side-chain dynamics. These data demonstrate that conformational dynamics and packing densities are not fully correlated in proteins and that encapsulation of cofactors by the polypeptide can provide a precisely structured environment resistant to change across a wide range of physical conditions.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


