用于预测 T 细胞受体-抗原-人类白细胞抗原结合的滑动-注意转换器神经结构
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
新抗原具有高特异性、低毒性和易于个性化等特点,可诱发免疫反应并清除癌细胞,是很有前途的免疫疗法靶点。然而,由于 T 细胞受体、抗原和人类白细胞抗原序列之间存在复杂的相互作用,识别有效的新抗原仍然十分困难。在这项研究中,我们将重要的物理和生物先验与变压器模型相结合,提出了物理启发滑动变压器(PISTE)。在 PISTE 中,传统的数据驱动注意力机制被物理驱动动力学所取代,物理驱动动力学引导氨基酸残基沿着其相互作用的梯度场进行定位。这样就能智能地浏览错综复杂的生物序列相互作用,从而提高 T 细胞受体-抗原-人类白细胞抗原结合预测的准确性,并对罕见序列进行稳健的泛化。此外,即使在没有三维结构训练数据的情况下,PISTE 也能有效地恢复残基级接触关系。我们将 PISTE 应用于多种免疫原性肿瘤类型,以确定新抗原并识别新抗原反应 T 细胞。在一项前瞻性前列腺癌研究中,75% 的患者通过 PISTE 预测的新抗原产生了免疫反应。本文章由计算机程序翻译,如有差异,请以英文原文为准。
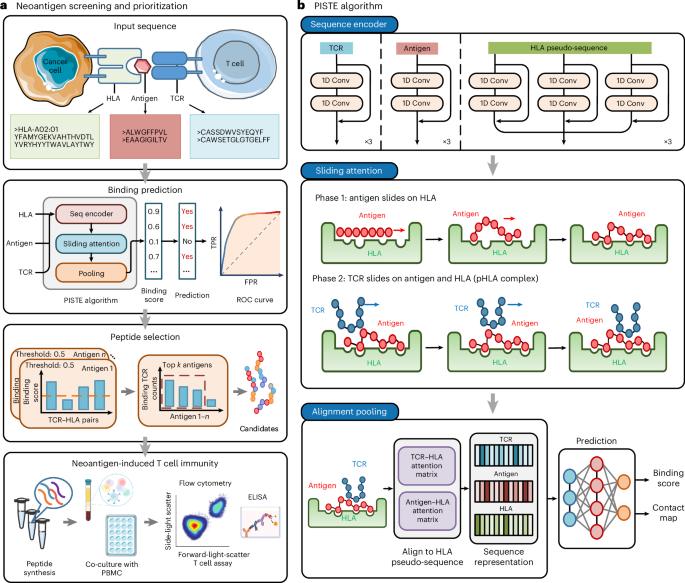
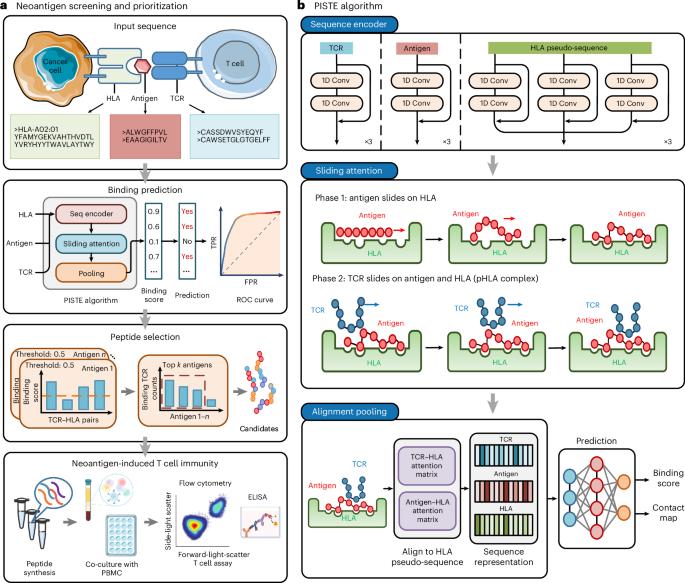
Sliding-attention transformer neural architecture for predicting T cell receptor–antigen–human leucocyte antigen binding
Neoantigens are promising targets for immunotherapy by eliciting immune response and removing cancer cells with high specificity, low toxicity and ease of personalization. However, identifying effective neoantigens remains difficult because of the complex interactions among T cell receptors, antigens and human leucocyte antigen sequences. In this study, we integrate important physical and biological priors with the Transformer model and propose the physics-inspired sliding transformer (PISTE). In PISTE, the conventional, data-driven attention mechanism is replaced with physics-driven dynamics that steers the positioning of amino acid residues along the gradient field of their interactions. This allows navigating the intricate landscape of biosequence interactions intelligently, leading to improved accuracy in T cell receptor–antigen–human leucocyte antigen binding prediction and robust generalization to rare sequences. Furthermore, PISTE effectively recovers residue-level contact relationships even in the absence of three-dimensional structure training data. We applied PISTE in a multitude of immunogenic tumour types to pinpoint neoantigens and discern neoantigen-reactive T cells. In a prospective study of prostate cancer, 75% of the patients elicited immune responses through PISTE-predicted neoantigens. Predicting TCR–antigen–human leucocyte antigen binding opens the door to neoantigen identification. In this study, a physics-inspired sliding transformer (PISTE) system is used to guide the positioning of amino acid residues along the gradient field of their interactions, boosting binding prediction accuracy.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


