绘制超越基因组的人类血浆蛋白质组生物影响图谱
IF 18.9
1区 医学
Q1 ENDOCRINOLOGY & METABOLISM
引用次数: 0
摘要
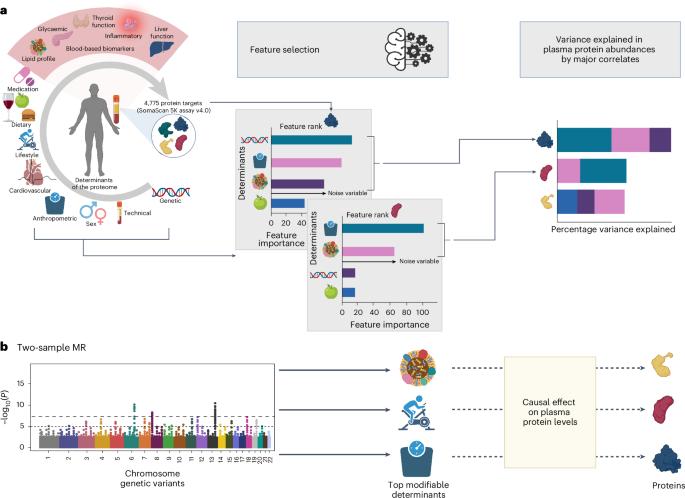
目前,广泛捕获的蛋白质组学平台可同时评估数千种血浆蛋白质,但其中大多数蛋白质并不是主动分泌的,其来源在很大程度上也是未知的。在这里,我们整合了基因组与深度表型组信息,在约 8,000 名大多数健康的个体中识别出与 4,775 种血浆蛋白相关的可调节和不可调节因素。我们绘制了一张数据驱动的人类血浆蛋白质组生物影响图,并展示了基于主要解释因素的蛋白质分群。对于超过三分之一(N = 1,575)的蛋白质目标,遗传和非遗传因素共同解释了血浆中 10-77% 的变化(中位数为 19.88%,四分位数间距为 14.01-31.09%),与技术因素无关(中位数为 2.48%,四分位数间距为 0.78-6.41%)。结合基因锚定因果推断方法,我们的图谱突出了可改变的风险因素与血浆蛋白之间的潜在因果关系,涉及数百种蛋白质与疾病的关联,例如 COL6A3,它可能介导了肾功能减退与心血管疾病之间的关联。我们提供了人类血浆蛋白质组的生物和技术影响图谱,以帮助人们理解蛋白质组研究结果的来龙去脉。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Mapping biological influences on the human plasma proteome beyond the genome
Broad-capture proteomic platforms now enable simultaneous assessment of thousands of plasma proteins, but most of these are not actively secreted and their origins are largely unknown. Here we integrate genomic with deep phenomic information to identify modifiable and non-modifiable factors associated with 4,775 plasma proteins in ~8,000 mostly healthy individuals. We create a data-driven map of biological influences on the human plasma proteome and demonstrate segregation of proteins into clusters based on major explanatory factors. For over a third (N = 1,575) of protein targets, joint genetic and non-genetic factors explain 10–77% of the variation in plasma (median 19.88%, interquartile range 14.01–31.09%), independent of technical factors (median 2.48%, interquartile range 0.78–6.41%). Together with genetically anchored causal inference methods, our map highlights potential causal associations between modifiable risk factors and plasma proteins for hundreds of protein–disease associations, for example, COL6A3, which possibly mediates the association between reduced kidney function and cardiovascular disease. We provide a map of biological and technical influences on the human plasma proteome to help contextualize findings from proteomic studies. The authors systematically study biological influences on the human plasma proteome in a large cohort, thereby revealing causal associations between plasma proteins and modifiable risk factors for protein–disease associations.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature metabolism
ENDOCRINOLOGY & METABOLISM-
CiteScore
27.50
自引率
2.40%
发文量
170
期刊介绍:
Nature Metabolism is a peer-reviewed scientific journal that covers a broad range of topics in metabolism research. It aims to advance the understanding of metabolic and homeostatic processes at a cellular and physiological level. The journal publishes research from various fields, including fundamental cell biology, basic biomedical and translational research, and integrative physiology. It focuses on how cellular metabolism affects cellular function, the physiology and homeostasis of organs and tissues, and the regulation of organismal energy homeostasis. It also investigates the molecular pathophysiology of metabolic diseases such as diabetes and obesity, as well as their treatment. Nature Metabolism follows the standards of other Nature-branded journals, with a dedicated team of professional editors, rigorous peer-review process, high standards of copy-editing and production, swift publication, and editorial independence. The journal has a high impact factor, has a certain influence in the international area, and is deeply concerned and cited by the majority of scholars.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


