燕麦多肽降血糖功能的虚拟筛选、分子对接和分子动力学模拟研究
IF 2.7
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
燕麦中含有大量的多糖和多肽,其中燕麦多肽具有降低体内血糖水平的功能。本文综述了从文献中提取和鉴定的燕麦多肽,并构建了相应的燕麦多肽数据库。以 DPP4 蛋白为基础,对肽数据库进行了虚拟筛选。对筛选得到的六种多肽进行了一百纳秒分子动力学模拟。根据模拟后的稳定结合构象分析了不同多肽分子与 DPP4 之间的相互作用信息,并计算了不同多肽分子与 DPP4 之间的结合自由能。结果表明,虚拟筛选得到的多肽分子均能与DPP4蛋白稳定结合,其中有两个多肽分子与DPP4的亲和力相对较强,可作为先导分子用于后续DPP4抑制剂的设计和修饰。模拟结果对深入理解 DPP4 的结构特征以及 DPP4 与燕麦肽的分子识别机制具有参考价值。本文章由计算机程序翻译,如有差异,请以英文原文为准。
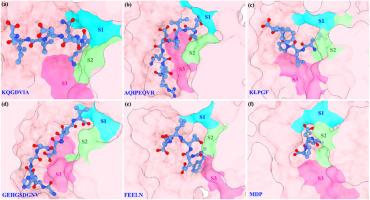
Virtual screening, molecular docking, and molecular dynamics simulation studies on the hypoglycemic function of oat peptides
Oat contains a large amount of polysaccharides and peptides, among which oat peptides have the function of reducing blood sugar levels in the body. This paper reviewed the peptides obtained from oat extraction and identification from literature and constructed the corresponding oat peptide database. Based on the DPP4 protein, a virtual screening of the peptide database was performed. One hundred nanosecond molecular dynamics simulations were performed for the six peptides obtained from the screening. The interaction information between different peptide molecules and DPP4 was analyzed from the stable binding conformations after the simulation, and the binding free energy between different peptide molecules and DPP4 was calculated. The results show that the peptide molecules obtained from the virtual screening can all stably bind to the DPP4 protein, among which two peptide molecules have relatively strong affinity with DPP4 and can be used as lead molecules for the subsequent design and modification of DPP4 inhibitors. The simulation results are informative for a deeper understanding of the structural characteristics of DPP4 and the molecular recognition mechanism between DPP4 and oat peptides.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of molecular graphics & modelling
生物-计算机:跨学科应用
CiteScore
5.50
自引率
6.90%
发文量
216
审稿时长
35 days
期刊介绍:
The Journal of Molecular Graphics and Modelling is devoted to the publication of papers on the uses of computers in theoretical investigations of molecular structure, function, interaction, and design. The scope of the journal includes all aspects of molecular modeling and computational chemistry, including, for instance, the study of molecular shape and properties, molecular simulations, protein and polymer engineering, drug design, materials design, structure-activity and structure-property relationships, database mining, and compound library design.
As a primary research journal, JMGM seeks to bring new knowledge to the attention of our readers. As such, submissions to the journal need to not only report results, but must draw conclusions and explore implications of the work presented. Authors are strongly encouraged to bear this in mind when preparing manuscripts. Routine applications of standard modelling approaches, providing only very limited new scientific insight, will not meet our criteria for publication. Reproducibility of reported calculations is an important issue. Wherever possible, we urge authors to enhance their papers with Supplementary Data, for example, in QSAR studies machine-readable versions of molecular datasets or in the development of new force-field parameters versions of the topology and force field parameter files. Routine applications of existing methods that do not lead to genuinely new insight will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


