傅立叶空间的选择性信号增强,作为从原位 TEM 发现大分子超微结构组织的工具。
IF 2.7
3区 生物学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
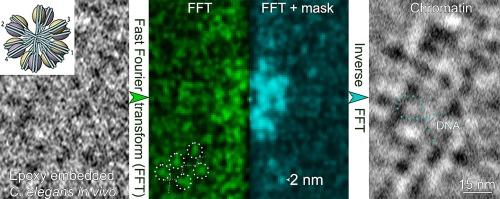
我们提出了一种基于傅立叶变换(FT)的分析方法,通过对奇异的大分子电子显微镜密度信息进行选择性过滤,可以从亚纳米尺度的 TEM 图像中获得超微结构细节。它适用于高压冷冻、冷冻水合和环氧树脂冷冻替代和嵌入的生物物种。二维投影和来自重建断层扫描的正切片都可用作结构信息的来源。该方法的关键在于选择感兴趣的大分子或细胞器,精确度≥ 7 - 3 nm(取决于初始倾斜序列或奇异图像采集的像素大小),并探索中心低频 FT 强度和衍射区域,以分别获得空间结构组织及其尺寸特征。我们还介绍了一种针对特定结构的选择性掩膜 FT 滤波方法,即使在对比度较差的树脂切片 TEM 中也能显著改善图像信息,而无需使用重金属。所述方法无需平均值即可阐明染色质结构。研究人员在体内发现了直径为 30 nm 的染色质纤维呈人字形对称,而这种染色质纤维在图像中保留了亚纳米级的细节。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Selective signal enhancement in Fourier space as a tool for discovering ultrastructural organization of macromolecules from in situ TEM
We present a Fourier transform (FT) based analytical method that allows to obtain of ultrastructural details from TEM images at sub-nanometer scale applying a selective filtering for singular macromolecule electron microscopy density information. It can be applied to high-pressure frozen, frozen hydrated and epoxy freeze substituted and embedded biological species. Both 2D projections and orthoslices from reconstructed tomograms can be used as a source of structural information. The key to the method is to select the macromolecule or organelle of interest with an accuracy of ≥ 7 – 3 nm (depending on pixel size of initial tilt series or singular image acquisition) and explore both the central low frequency FT intensity and diffraction regions to obtain the spatial structural organization and its dimensional characteristics, respectively. We also introduce a structure-specific selective mask FT filtering approach that can significantly improve image information even in poorly contrasted TEM of resin sections without heavy metal been used. The described method elucidates chromatin architecture without the need of averaging. A zigzag symmetry of 30 nm diameter chromatin fibers which in general is a controversial topic of research has been identified for C. elegans cells in vivo with sub-nanometer details being preserved in the images.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of structural biology
生物-生化与分子生物学
CiteScore
6.30
自引率
3.30%
发文量
88
审稿时长
65 days
期刊介绍:
Journal of Structural Biology (JSB) has an open access mirror journal, the Journal of Structural Biology: X (JSBX), sharing the same aims and scope, editorial team, submission system and rigorous peer review. Since both journals share the same editorial system, you may submit your manuscript via either journal homepage. You will be prompted during submission (and revision) to choose in which to publish your article. The editors and reviewers are not aware of the choice you made until the article has been published online. JSB and JSBX publish papers dealing with the structural analysis of living material at every level of organization by all methods that lead to an understanding of biological function in terms of molecular and supermolecular structure.
Techniques covered include:
• Light microscopy including confocal microscopy
• All types of electron microscopy
• X-ray diffraction
• Nuclear magnetic resonance
• Scanning force microscopy, scanning probe microscopy, and tunneling microscopy
• Digital image processing
• Computational insights into structure
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


