紫木匠蜂(Xylocopa violacea,林尼厄斯,1785 年)的基因组序列:一个正在扩大范围的膜翅目物种
IF 3.1
2区 生物学
Q2 ECOLOGY
引用次数: 0
摘要
我们展示了一个来自雄性紫木匠蜂(Xylocopa violacea, Linnaeus 1758)个体的参考基因组组装。该基因组的跨度为 1.02 千亿字节。48%的基因组组装为 17 个假染色体单元。线粒体基因组也已组装完成,长度为 21.8 千碱基。该基因组具有高度重复性,可能代表了 Xylocopa 属蜜蜂的高度异染色体结构。我们还使用基于证据的方法注释了 10,152 个高置信度编码基因。该基因组的测序是欧洲参考基因组图谱(ERGA)试点项目的一部分,是对膜翅目昆虫基因组资源的重要补充。本文章由计算机程序翻译,如有差异,请以英文原文为准。
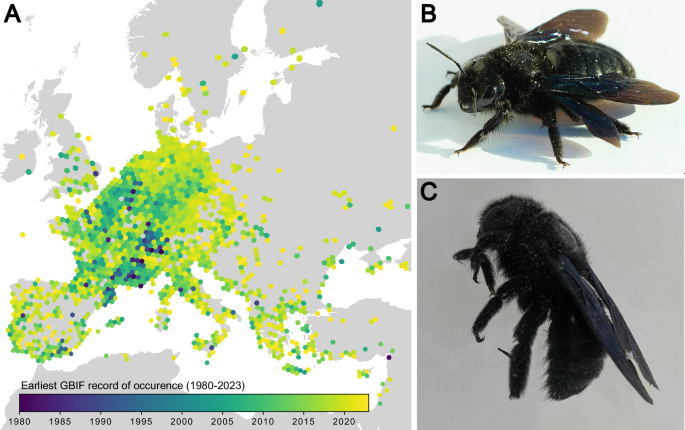
The genome sequence of the Violet Carpenter Bee, Xylocopa violacea (Linnaeus, 1785): a hymenopteran species undergoing range expansion
We present a reference genome assembly from an individual male Violet Carpenter Bee (Xylocopa violacea, Linnaeus 1758). The assembly is 1.02 gigabases in span. 48% of the assembly is scaffolded into 17 pseudo-chromosomal units. The mitochondrial genome has also been assembled and is 21.8 kilobases in length. The genome is highly repetitive, likely representing a highly heterochromatic architecture expected of bees from the genus Xylocopa. We also use an evidence-based methodology to annotate 10,152 high confidence coding genes. This genome was sequenced as part of the pilot project of the European Reference Genome Atlas (ERGA) and represents an important addition to the genomic resources available for Hymenoptera.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Heredity
生物-进化生物学
CiteScore
7.50
自引率
2.60%
发文量
84
审稿时长
4-8 weeks
期刊介绍:
Heredity is the official journal of the Genetics Society. It covers a broad range of topics within the field of genetics and therefore papers must address conceptual or applied issues of interest to the journal''s wide readership
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


