从溶液和固态核磁共振深入了解配体介导的寡聚环形基因调控蛋白的活化过程
IF 4.7
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
91 kDa 的寡聚环形配体结合蛋白 TRAP(trp RNA 结合衰减蛋白)调节着一系列参与色氨酸(Trp)在芽孢杆菌中生物合成的基因的表达。当细胞中的 Trp 水平升高时,游离氨基酸就会与埋藏在环中 11 个(或 12 个,视物种而定)原生体之间界面上的位点结合。与 Trp 结合的 TRAP 的晶体结构显示,Trp 配体通过相邻原体的一对环路与溶剂隔离,这对环路通过与几个苏氨酸残基的极性接触将结合的配体埋藏起来。Trp 配体的结合是协同进行的,因此连续的结合事件会以更高的表观亲和力发生,但这种协同性的结构基础却鲜为人知。我们利用以苏氨酸和异亮氨酸侧链为重点的溶液甲基-TROSY NMR 驰豫实验,以及以 800 和 1200 MHz 频率记录的均匀标记样品的魔角旋转固态 NMR 13C-13C 和 15N-13C 化学位移相关光谱,来描述该蛋白质的结构和动力学特征。对不含配体的 apo TRAP 进行的甲基 13C 驰豫弥散实验显示,在 µs-ms 时间尺度上存在一致的交换动力学,这与瞬时取样构象可能允许配体结合是一致的。交叉相关弛豫实验揭示了快速时间尺度上的广泛无序性。在载脂蛋白和 Trp 结合的 TRAP 中,含甲基侧链的化学位移显示了取样侧链旋转态分布的微妙变化。这些观察结果揭示了诱导构象变化以产生同向 Trp-Trp 结合合作性的途径和机制。本文章由计算机程序翻译,如有差异,请以英文原文为准。
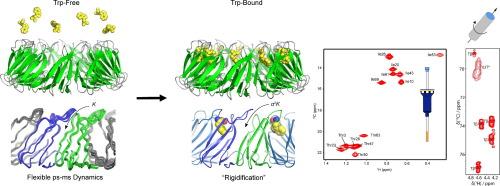
Insights into Ligand-Mediated Activation of an Oligomeric Ring-Shaped Gene-Regulatory Protein from Solution- and Solid-State NMR
The 91 kDa oligomeric ring-shaped ligand binding protein TRAP (trp RNA binding attenuation protein) regulates the expression of a series of genes involved in tryptophan (Trp) biosynthesis in bacilli. When cellular Trp levels rise, the free amino acid binds to sites buried in the interfaces between each of the 11 (or 12, depending on the species) protomers in the ring. Crystal structures of Trp-bound TRAP show the Trp ligands are sequestered from solvent by a pair of loops from adjacent protomers that bury the bound ligand via polar contacts to several threonine residues. Binding of the Trp ligands occurs cooperatively, such that successive binding events occur with higher apparent affinity but the structural basis for this cooperativity is poorly understood. We used solution methyl-TROSY NMR relaxation experiments focused on threonine and isoleucine sidechains, as well as magic angle spinning solid-state NMR 13C–13C and 15N-13C chemical shift correlation spectra on uniformly labeled samples recorded at 800 and 1200 MHz, to characterize the structure and dynamics of the protein. Methyl 13C relaxation dispersion experiments on ligand-free apo TRAP revealed concerted exchange dynamics on the µs-ms time scale, consistent with transient sampling of conformations that could allow ligand binding. Cross-correlated relaxation experiments revealed widespread disorder on fast timescales. Chemical shifts for methyl-bearing side chains in apo- and Trp-bound TRAP revealed subtle changes in the distribution of sampled sidechain rotameric states. These observations reveal a pathway and mechanism for induced conformational changes to generate homotropic Trp-Trp binding cooperativity.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


