用于钙成像数据去噪、检测和去混合的端到端循环压缩传感方法
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
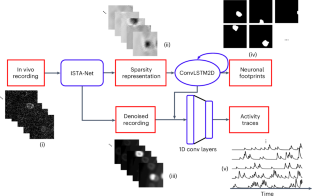
双光子钙成像技术能以细胞分辨率对神经元活动进行大规模记录。在分析钙成像数据时,我们非常需要一种稳健、自动化和高速的管道来同时分割神经元的空间足迹并提取其时间活动轨迹,同时消除背景、噪声和重叠神经元的污染。在这里,我们展示了 DeepCaImX,一种基于迭代收缩阈值算法和长短期记忆神经网络的端到端深度学习方法,它能以极高的速度实现上述目标,而且无需手动调整超参数。DeepCaImX 是一种多任务、多类和多标签分割方法,由一个包含递归层和全连接层的压缩传感启发神经网络组成。该神经网络可同时生成精确的神经元足迹,并从钙成像数据中提取干净的神经元活动轨迹。我们用模拟数据集训练了神经网络,并用体内实验数据将其与现有的最先进方法进行了比较。DeepCaImX 在分割和时间轨迹提取质量以及处理速度方面都优于现有方法。DeepCaImX 具有很强的可扩展性,对中尺度钙成像分析大有裨益。本文章由计算机程序翻译,如有差异,请以英文原文为准。

An end-to-end recurrent compressed sensing method to denoise, detect and demix calcium imaging data
Two-photon calcium imaging provides large-scale recordings of neuronal activities at cellular resolution. A robust, automated and high-speed pipeline to simultaneously segment the spatial footprints of neurons and extract their temporal activity traces while decontaminating them from background, noise and overlapping neurons is highly desirable to analyse calcium imaging data. Here we demonstrate DeepCaImX, an end-to-end deep learning method based on an iterative shrinkage-thresholding algorithm and a long short-term memory neural network to achieve the above goals altogether at a very high speed and without any manually tuned hyperparameter. DeepCaImX is a multi-task, multi-class and multi-label segmentation method composed of a compressed sensing-inspired neural network with a recurrent layer and fully connected layers. The neural network can simultaneously generate accurate neuronal footprints and extract clean neuronal activity traces from calcium imaging data. We trained the neural network with simulated datasets and benchmarked it against existing state-of-the-art methods with in vivo experimental data. DeepCaImX outperforms existing methods in the quality of segmentation and temporal trace extraction as well as processing speed. DeepCaImX is highly scalable and will benefit the analysis of mesoscale calcium imaging. Extracting time traces and spatial footprints of single neurons from population calcium imaging data presents challenges. Zhang et al. introduce a deep learning method that efficiently segments neuronal footprints and extracts activity traces from these data. The method surpasses existing approaches in both quality and speed, providing a robust tool for large-scale neuronal circuit analysis.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


