利用分数去噪进行预训练,提高分子特性预测能力
IF 23.9
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
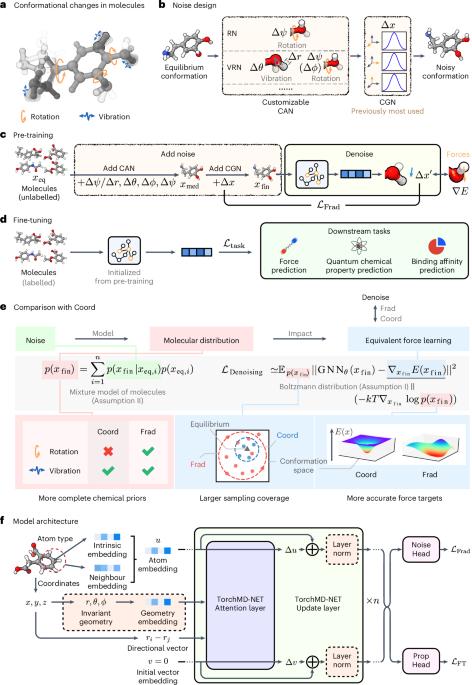
深度学习方法被认为有望加速药物发现和材料设计中的分子筛选。由于标记数据的可用性有限,人们提出了各种自监督分子预训练方法。虽然许多现有方法利用了计算机视觉和自然语言处理中常见的预训练任务,但它们往往忽略了分子的基本物理原理。相比之下,在预训练中应用去噪可以解释为等效的力学习,但有限的噪声分布会给分子分布带来偏差。为了解决这个问题,我们引入了一种称为分数去噪的分子预训练框架,它将噪声设计与力学习等效性所施加的约束分离开来。通过这种方式,噪声变得可定制,从而可以结合化学先验,大幅改进分子分布建模。实验证明,我们的框架始终优于现有方法,在力预测、量子化学特性和结合亲和力任务方面取得了最先进的结果。经过改进的噪声设计提高了力的准确性和采样覆盖率,有助于创建物理上一致的分子表征,最终实现卓越的预测性能。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Pre-training with fractional denoising to enhance molecular property prediction
Deep learning methods have been considered promising for accelerating molecular screening in drug discovery and material design. Due to the limited availability of labelled data, various self-supervised molecular pre-training methods have been presented. Although many existing methods utilize common pre-training tasks in computer vision and natural language processing, they often overlook the fundamental physical principles governing molecules. In contrast, applying denoising in pre-training can be interpreted as an equivalent force learning, but the limited noise distribution introduces bias into the molecular distribution. To address this issue, we introduce a molecular pre-training framework called fractional denoising, which decouples noise design from the constraints imposed by force learning equivalence. In this way, the noise becomes customizable, allowing for incorporating chemical priors to substantially improve the molecular distribution modelling. Experiments demonstrate that our framework consistently outperforms existing methods, establishing state-of-the-art results across force prediction, quantum chemical properties and binding affinity tasks. The refined noise design enhances force accuracy and sampling coverage, which contribute to the creation of physically consistent molecular representations, ultimately leading to superior predictive performance. Denoising methods introduce useful priors in pre-training methods for molecular property prediction, but chemically unaware noise can lead to inaccurate predictions in downstream tasks. A molecular pre-training framework that uses fractional denoising to improve molecular distribution modelling is proposed, resulting in better predictions in various property prediction tasks.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


