保留 ES 细胞衍生的 129S 基因组会导致 Nlrp3tm1Flv 小鼠 NLRP1 超敏和转录失调
IF 13.7
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
人类和小鼠的免疫反应基因具有高度的多态性,不同基因座之间的异质性驱动着菌株特异性的宿主防御反应。无意中保留的多态性位点会带来混杂的表型,导致错误的结论,阻碍科学进步。在这项研究中,我们结合使用了 RNAseq 和变异调用分析,在最常用的 NLRP3 缺乏症小鼠模型之一(Nlrp3tm1Flv)中鉴定了 129S 基因组的一个重要区域,包括 Nlrp3 近端高度多态的 Nlrp1 基因座。我们的研究表明,Nlrp1129S 基因座的存在导致 NLRP1B 蛋白表达增加,并使 Nlrp3tm1Flv 巨噬细胞对 NLRP1 炎症小体的激活敏感,而与 NLRP3 缺乏无关。129S 基因组的保留进一步导致蛋白质序列差异和多种细胞类型基因调控的改变,包括关键的组织驻留巨噬细胞标记物 TIM4。利用 NLRP3 缺乏症的替代模型,包括以前未曾描述过的条件性 Nlrp3 等位基因,对 Nlrp3 缺失进行精确的时间和细胞类型特异性控制,我们进一步表明 NLRP3 对 Talabostat 驱动的 IL-1β 释放做出了贡献。我们的研究还建立了一个通用框架,利用 RNAseq 数据鉴定功能相关的 SNPs 并评估转基因小鼠的基因组污染。这样就能将表型明确归因于目标基因,提高宿主防御反应领域研究的精确性和可靠性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
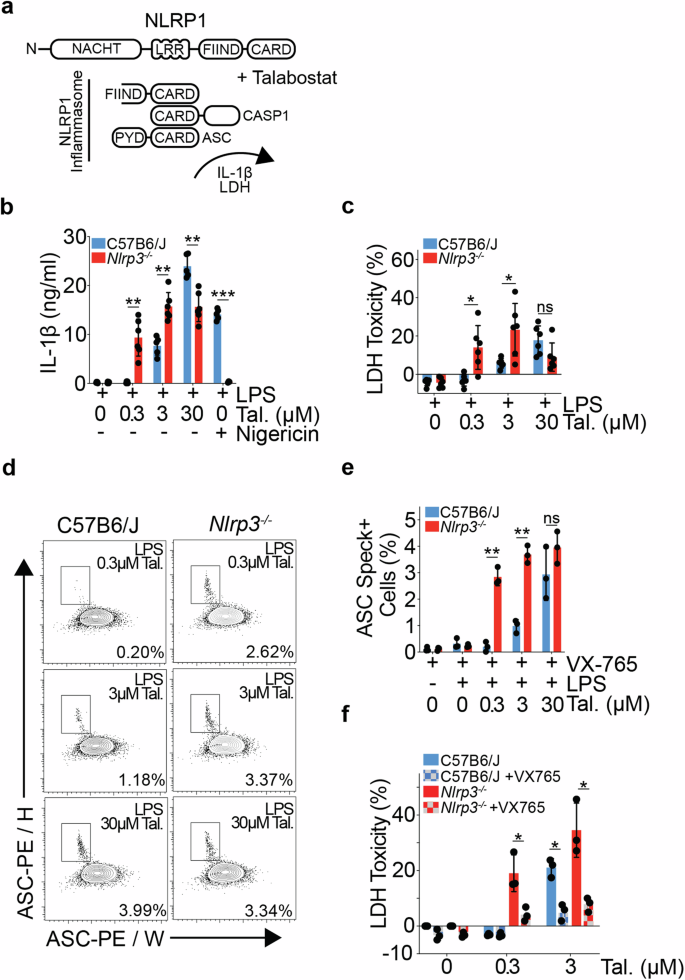
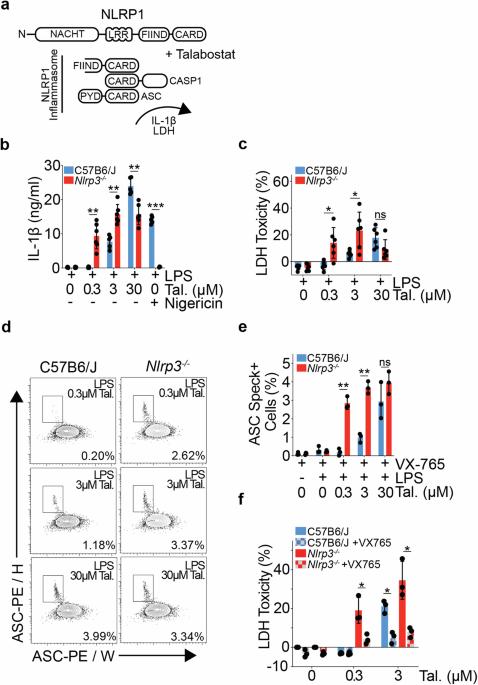
Retention of ES cell-derived 129S genome drives NLRP1 hypersensitivity and transcriptional deregulation in Nlrp3tm1Flv mice
Immune response genes are highly polymorphic in humans and mice, with heterogeneity amongst loci driving strain-specific host defence responses. The inadvertent retention of polymorphic loci can introduce confounding phenotypes, leading to erroneous conclusions, and impeding scientific advancement. In this study, we employ a combination of RNAseq and variant calling analyses to identify a substantial region of 129S genome, including the highly polymorphic Nlrp1 locus, proximal to Nlrp3, in one of the most commonly used mouse models of NLRP3 deficiency (Nlrp3tm1Flv). We show that the presence of the Nlrp1129S locus leads to an increase in NLRP1B protein expression, and a sensitising of Nlrp3tm1Flv macrophages to NLRP1 inflammasome activation, independent of NLRP3 deficiency. Retention of 129S genome further leads to protein sequence differences and altered gene regulation across multiple cell types, including of the key tissue-resident macrophage marker, TIM4. Using alternative models of NLRP3 deficiency, including a previously undescribed conditional Nlrp3 allele enabling precise temporal and cell-type specific control over Nlrp3 deletion, we further show that NLRP3 contributes to Talabostat-driven IL-1β release. Our study also establishes a generic framework to identify functionally relevant SNPs and assess genomic contamination in transgenic mice using RNAseq data. This allows for unambiguous attribution of phenotypes to the target gene and advances the precision and reliability of research in the field of host defence responses.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell Death and Differentiation
生物-生化与分子生物学
CiteScore
24.70
自引率
1.60%
发文量
181
审稿时长
3 months
期刊介绍:
Mission, vision and values of Cell Death & Differentiation:
To devote itself to scientific excellence in the field of cell biology, molecular biology, and biochemistry of cell death and disease.
To provide a unified forum for scientists and clinical researchers
It is committed to the rapid publication of high quality original papers relating to these subjects, together with topical, usually solicited, reviews, meeting reports, editorial correspondence and occasional commentaries on controversial and scientifically informative issues.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


